Sequence analysis of 2qgn: Difference between revisions
From MDWiki
Jump to navigationJump to search
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
==Multiple Sequence Alignment== | |||
[[Image:RR1.png|framed|'''(A)''']] | |||
[[Image:RR2.png|framed|'''(B)''']] | |||
[[Image:RR3.png|framed|'''(C)''']] | |||
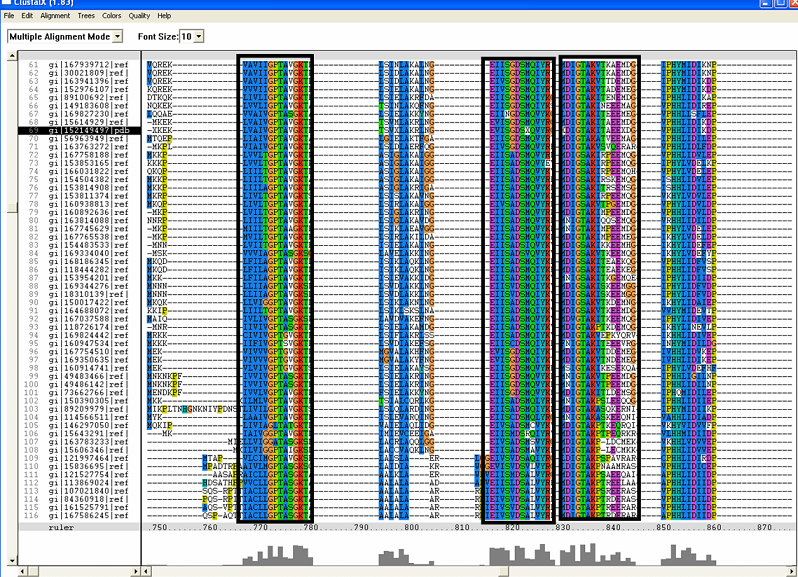
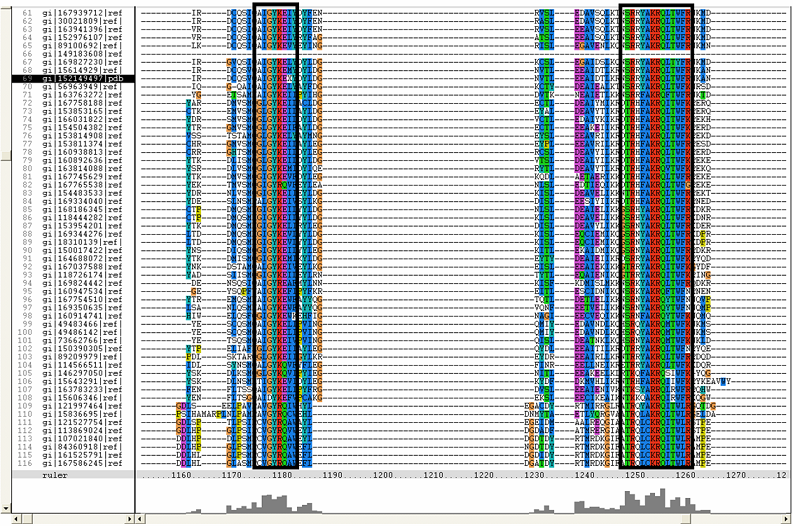
[[Image:ZZZ.png|framed|'''(D)'''<P><B>Figure 1</B> : <BR>Multiple sequence alignment with 260 homologous sequences and 2qgnA (tRNA isopentenyltransferase 1, 69th sequence which is highlighted on the right hand column) was constructed by ClustalX. Gaps are represented as ‘-‘. Orange, red, blue and green indicate residue code of “A”, “C”, “T” and “G” respectively (Kohli and Bachhawat 2003). Conserved regions are shown in the black box. <B>(A)</B> region from 750bp to 870 bps <B>(B)</B> region from 880bp to 1000bp <B>(C)</B> region from 1160bp to 1270bp <B>(D)</B>region on the c-terminus.|none]] | |||
[[FIGTREE|next]] | |||
Revision as of 12:51, 9 June 2008
Multiple Sequence Alignment
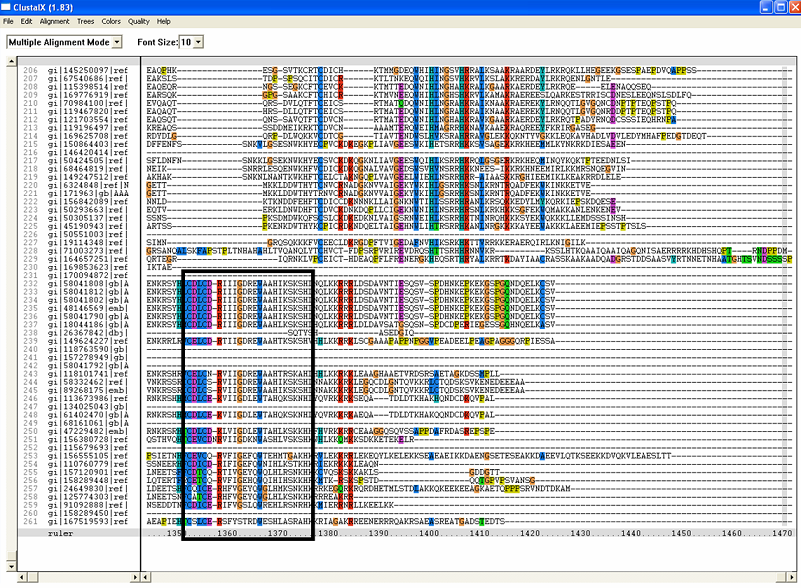
(D)
Figure 1 :
Multiple sequence alignment with 260 homologous sequences and 2qgnA (tRNA isopentenyltransferase 1, 69th sequence which is highlighted on the right hand column) was constructed by ClustalX. Gaps are represented as ‘-‘. Orange, red, blue and green indicate residue code of “A”, “C”, “T” and “G” respectively (Kohli and Bachhawat 2003). Conserved regions are shown in the black box. (A) region from 750bp to 870 bps (B) region from 880bp to 1000bp (C) region from 1160bp to 1270bp (D)region on the c-terminus.