Main Hypothetical Protein Function Page: Difference between revisions
No edit summary |
|||
| (17 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
=='''Literature review'''== | |||
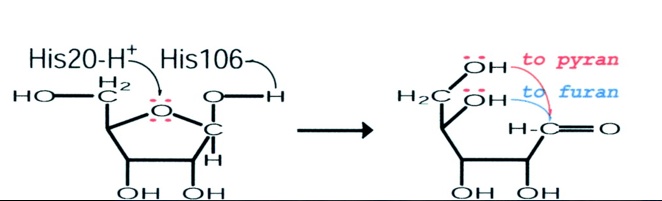
[[Image:rbsfunction.jpg]] | |||
(Ryu, et al., 2004) | |||
This schematic representation shows a proposed mode of action of the RbsD mutarotase. | |||
The articles findings lead to a proposed role for RbsD mutarotase. The His20 residue protonates the O5 and the His106 deprotonates the OH attached to the C1. This causes ring-breakage thus forming an equilibrium between the furan and pyran forms. | |||
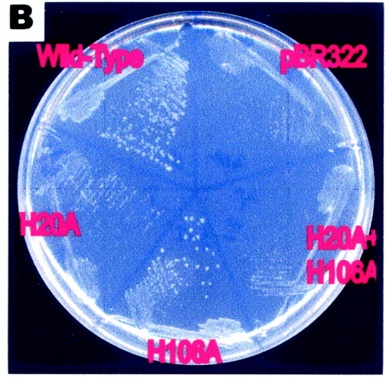
[[Image:petri.jpg]] | |||
(Ryu, et al., 2004) | |||
This image supports the proposed importance of the His residues in its pyranase activity. The growth of the e.coli represents rbsD activity. The H20A change completely abolished the pyranase activity, whereas H106A retained one-third of the original activity | |||
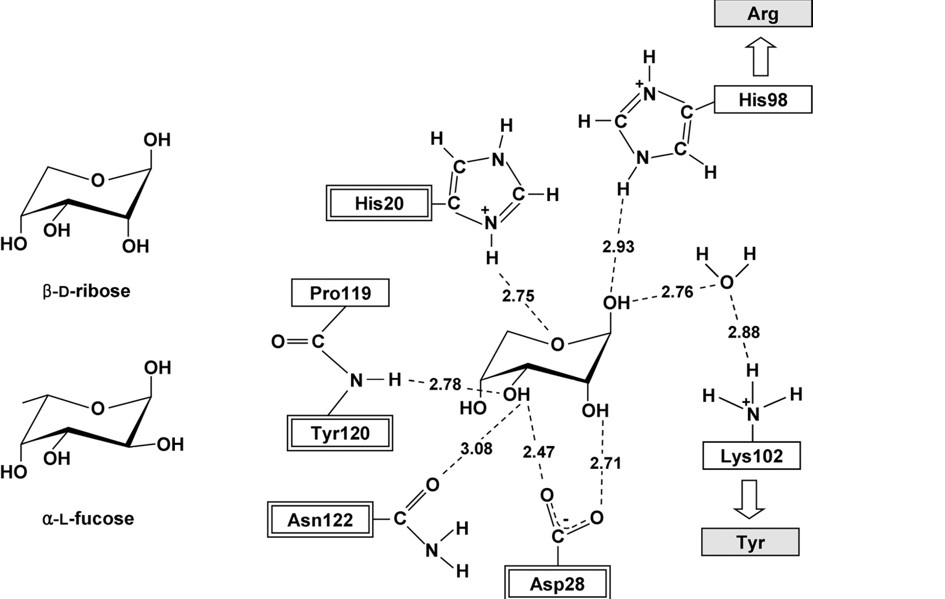
[[Image:Binding.jpg]] | |||
This schematic representation shows important residues that play a role in the binding of the sugar. H20, D28, Y120 and N122 are highly conserved residues in both RbsD and FucU proteins. The H98 and K102 are found in the RbsD protein but are exchanged for R and Y respectively for FucU proteins. The target protein has the R and Y residues leading to the conclusion that it is incolved with the conversion of the fucose sugar. | |||
=='''String Analysis'''== | =='''String Analysis'''== | ||
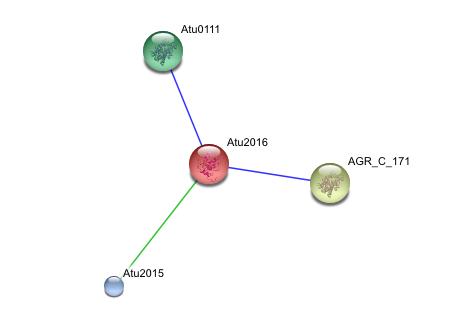
[[Image:String.jpg]] | |||
This string analysis shows possible protein - protein interactions. The red protein in the middle represents the target protein and the three surrounding proteins represent interacting proteins. | |||
The larger yellow and green proteins may be releated due to cooccurrence, where as the smaller blue protien may be related due to repeatedly occurring in close proximity to the target protein, however the posible interactions shown in the analysis are inconclusive and unsubstantiated. experimental evidence is needed to prove such relationships<br> | |||
=='''Profunc'''== | |||
===Interpro Scan result=== | |||
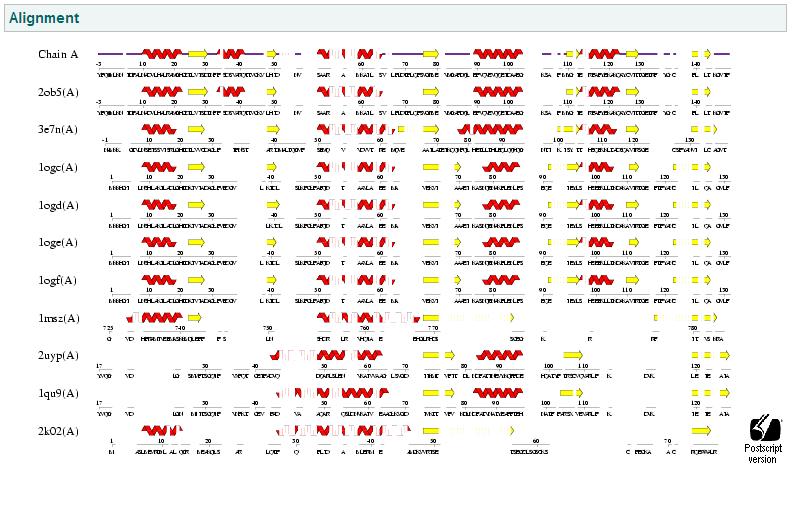
[[Image:Interpro1.gif]] | |||
This image shows the results of a InterProScan. The InterProScan search identifies sequence motifs from several databases.These results show that the target protein belongs to the RbsD/FucU superfamily (Purple and red lines) | |||
The gray line shows an possible binding histadine site. | |||
===PDB Sequence Match=== | |||
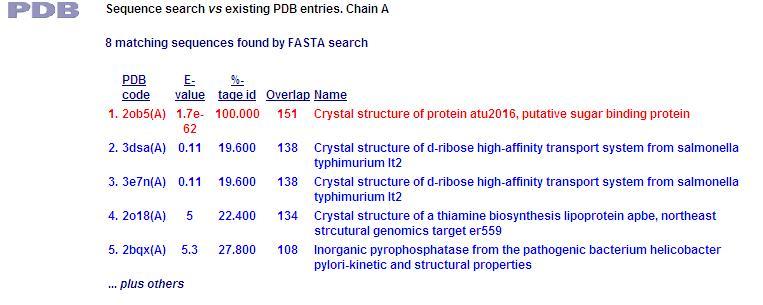
[[Image:PDB1.jpg]] | |||
This PDB scan compares structure alignment. The highest structural match with our sequence is shown to be involved in an d-ribose high affinity transport system. This supports the theory that our hypothetical protein is involved in the ABC transport system of ribose in prokaryote cells. | |||
===Residue Conservation Analysis=== | |||
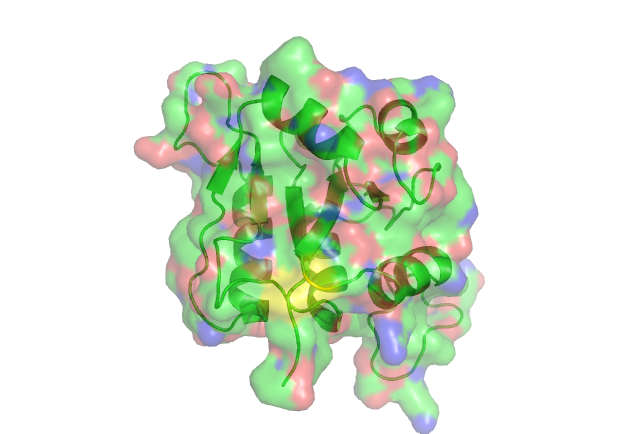
[[Image:pymol.png]] | |||
In this anaysis the residues are highlighted in different colours representing the levels of conservity. The red residues are the most conserved followed by pink, orange ect. <BR> | |||
The yellow residue is highly conserved. acourding to certain literature it is very functionally important in the related ribose_fucu superfamily. | |||
===Fold Match=== | |||
[[Image:FoldMatch.jpg]] | |||
This anaysis compares the similiar folds between structures. This again shows that the same d-ribose high affinity transport from the previous PDB analysis has the highest similarity, thus again reinforces the close relationship of our protein with ribose transport systems in prokaryote bacteria. This suggests similar function. | |||
[[hypothetical protein LOC282969 isoform 1 | Go Back To Main Page]] | [[hypothetical protein LOC282969 isoform 1 | Go Back To Main Page]] | ||
Latest revision as of 09:14, 9 June 2009
Literature review
This schematic representation shows a proposed mode of action of the RbsD mutarotase.
The articles findings lead to a proposed role for RbsD mutarotase. The His20 residue protonates the O5 and the His106 deprotonates the OH attached to the C1. This causes ring-breakage thus forming an equilibrium between the furan and pyran forms.
This image supports the proposed importance of the His residues in its pyranase activity. The growth of the e.coli represents rbsD activity. The H20A change completely abolished the pyranase activity, whereas H106A retained one-third of the original activity
This schematic representation shows important residues that play a role in the binding of the sugar. H20, D28, Y120 and N122 are highly conserved residues in both RbsD and FucU proteins. The H98 and K102 are found in the RbsD protein but are exchanged for R and Y respectively for FucU proteins. The target protein has the R and Y residues leading to the conclusion that it is incolved with the conversion of the fucose sugar.
String Analysis
This string analysis shows possible protein - protein interactions. The red protein in the middle represents the target protein and the three surrounding proteins represent interacting proteins.
The larger yellow and green proteins may be releated due to cooccurrence, where as the smaller blue protien may be related due to repeatedly occurring in close proximity to the target protein, however the posible interactions shown in the analysis are inconclusive and unsubstantiated. experimental evidence is needed to prove such relationships
Profunc
Interpro Scan result
This image shows the results of a InterProScan. The InterProScan search identifies sequence motifs from several databases.These results show that the target protein belongs to the RbsD/FucU superfamily (Purple and red lines) The gray line shows an possible binding histadine site.
PDB Sequence Match
This PDB scan compares structure alignment. The highest structural match with our sequence is shown to be involved in an d-ribose high affinity transport system. This supports the theory that our hypothetical protein is involved in the ABC transport system of ribose in prokaryote cells.
Residue Conservation Analysis
In this anaysis the residues are highlighted in different colours representing the levels of conservity. The red residues are the most conserved followed by pink, orange ect.
The yellow residue is highly conserved. acourding to certain literature it is very functionally important in the related ribose_fucu superfamily.
Fold Match
This anaysis compares the similiar folds between structures. This again shows that the same d-ribose high affinity transport from the previous PDB analysis has the highest similarity, thus again reinforces the close relationship of our protein with ribose transport systems in prokaryote bacteria. This suggests similar function.