Protein Evolution: Difference between revisions
No edit summary |
No edit summary |
||
| (21 intermediate revisions by the same user not shown) | |||
| Line 8: | Line 8: | ||
'''SUMMARY''' | '''SUMMARY''' | ||
08/05/2007 | |||
The protein sequence was transformed into FASTA format | The protein sequence was transformed into FASTA format | ||
| Line 26: | Line 28: | ||
[[Image:Ids.txt]] | [[Image:Ids.txt]] | ||
File that shows protein results | |||
[[Image:Protein-results.txt]] | |||
| Line 34: | Line 41: | ||
''Phylogenetic Tree'' | ''Phylogenetic Tree'' | ||
The first step was to perform a distance matrix calculation on the sequences. This was done by using the Phylip program on the CD. | The first step was to perform a distance matrix calculation on the sequences. This was done by using the Phylip program on the CD. Because the program only scans the first ten characters of the Id numbers of the sequences, several sequences had to be change because they appeared to be the same. These sequences were sequences 21 & 22, 23 & 24, 32 & 33, 39 & 40 & 41 & 47, and to be on the safe side 42 & 43 need to be changed also. | ||
22/05/2007 | |||
Original sequences and change to the sequences | |||
21 - gi|114673074 | |||
22 - gi|114673072 | |||
21 - kept the same | |||
22 - changed to gi|1146jpe | |||
23 - gi|109122125 | |||
24 - gi|109122123 | |||
23 - kept the same | |||
24 - changed to gi|1091jpe | |||
32 - gi|123258675 | |||
33 - gi|123258677 | |||
32 - kept the same | |||
33 - changed to gi|1232jpe | |||
39 - gi|119609670 | |||
40 - gi|119609672 | |||
41 - gi|119609671 | |||
47 - gi|119609669 | |||
39 - kept the same | |||
40 - changed to gi|1196jpe | |||
41 - changed to gi|1196epj | |||
47 - changed to gi|1196pje | |||
42 - gi|73964908 | |||
43 - gi|73964910 | |||
42 - kept the same | |||
43 - changed to gi|7396jpe | |||
Redid the distance matrix calculation with the newly changed sequences. This produced a file of the calculations which was called outputsequences.phy. (I can't upload .phy files onto the page) | |||
The first tree constructed was done using the neighbour-joining method. The output file was named neighbourjoiningspeciessequences.phy and the other output file that was produced from this method was called treealignment.ph The neighbour joining file was converted to a text file. | |||
File that shows the neighbour joining method which produced an unrooted tree | |||
[[Image:neighbourjoiningspeciessequences.txt]] | |||
This data was used to convert the tree into an image by using the Phylip program. | |||
(Still unable to upload phylogenetic tree, it is a work in progress) | |||
When the tree was completed, bootstrapping of the tree was the next task undertaken. This was done by following the instructions on the Methods section and using the Phylogenetic tree instructions. Part A bootstrapped the sequences 100 times. Part B was producing the bootstrap distance matrices. Step C was taking this file of bootstrap values and creating a tree alingment with these values using the programme neighbor. | |||
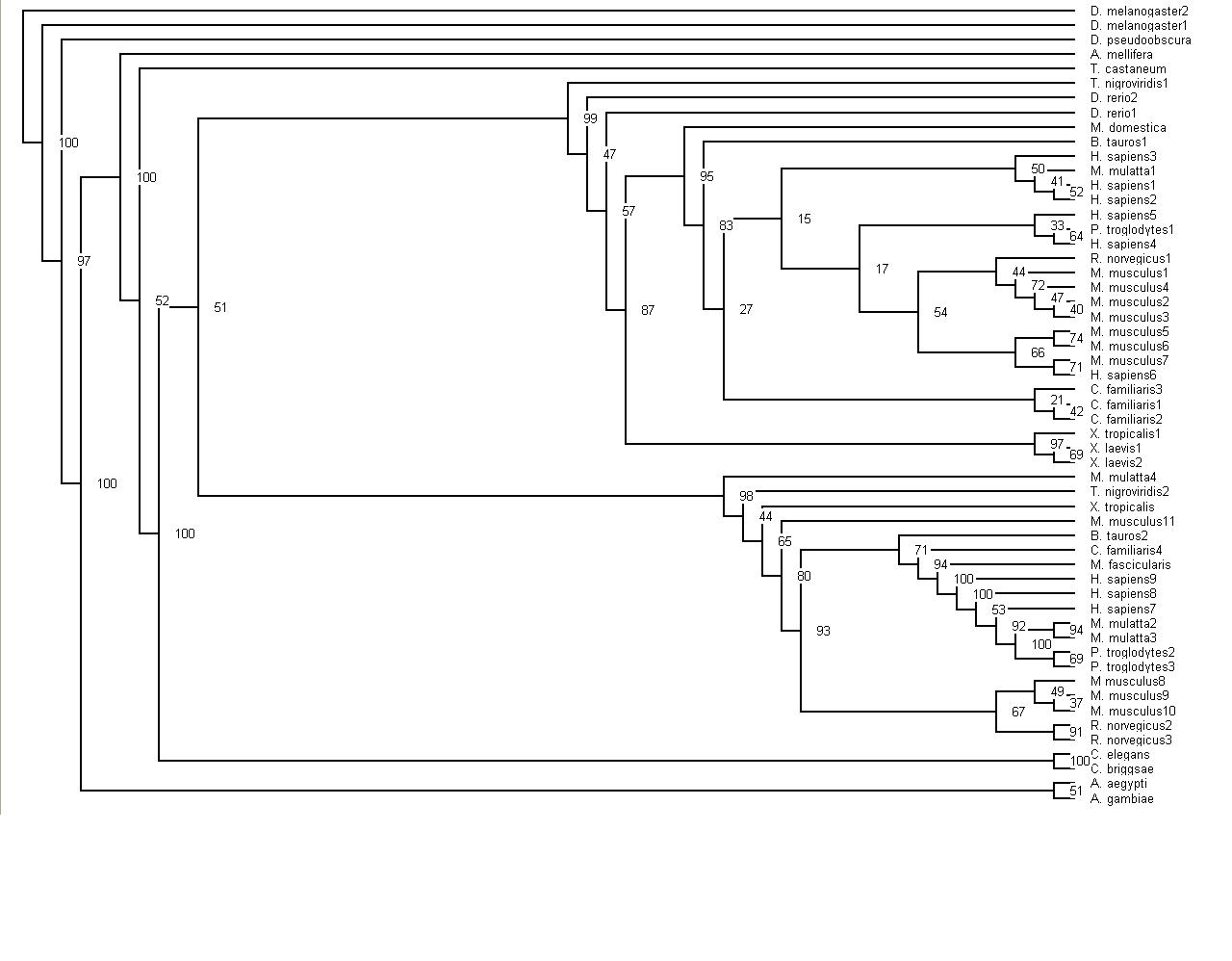
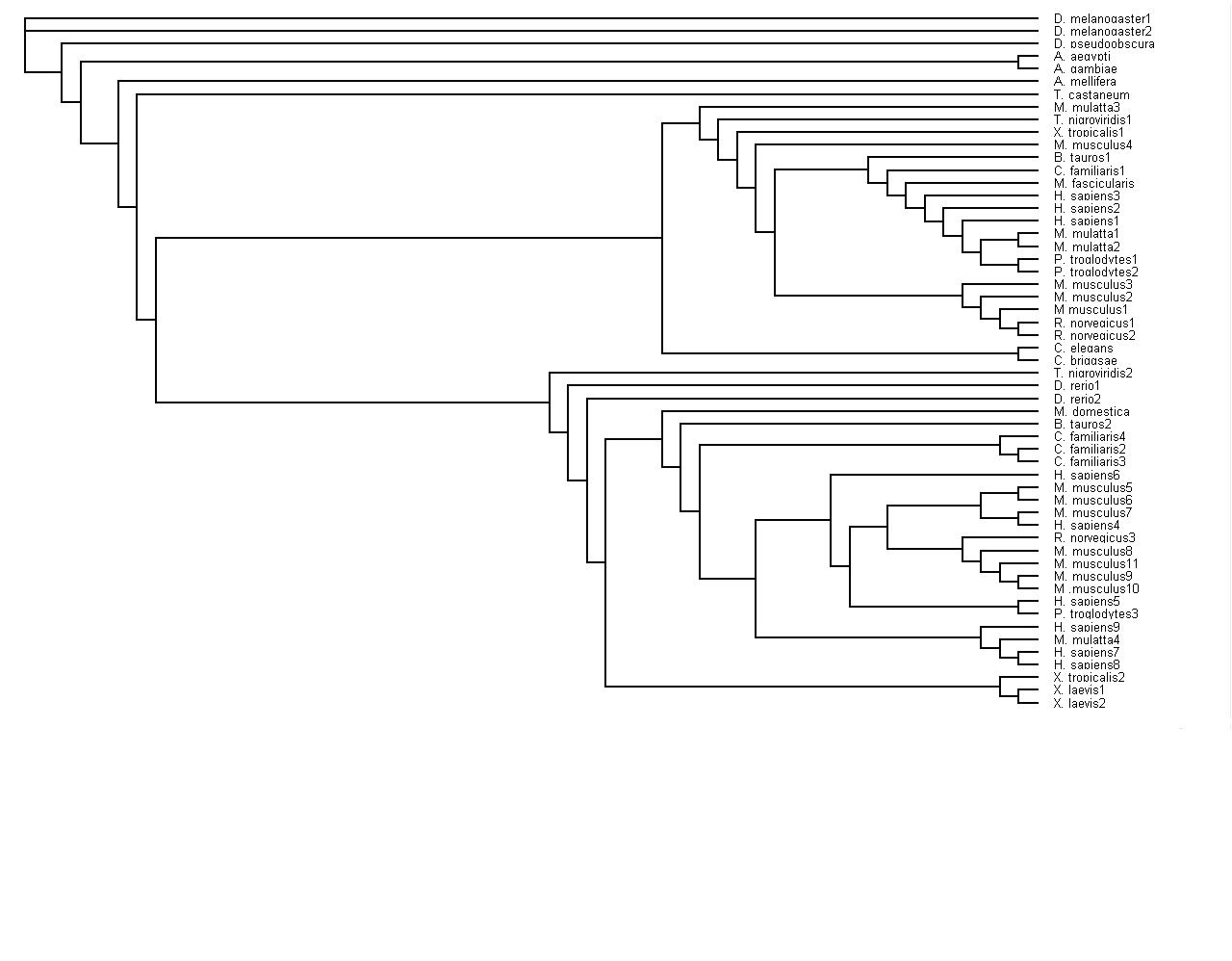
Finally, have been able to upload two tree diagrams that are slightly different, one has the bootstrap values | |||
[[Image:Bootstrap_Tree_Alignment.jpg]] | |||
[[Image:Tree_Alignment.jpg]] | |||
Access to [[Scientific Report]] | |||
Latest revision as of 04:59, 5 June 2007
Conservation of MIF4GD in Eutheria (Placental Mammals)
Conservation of the protein structure in many organisms, including Pan troglodytes (chimpanzee), Mus musculus (house mouse), Rattus norvegicus (brown rat), Bos taurus (domestic cow) and Danio rerio (zebrafish).
SUMMARY
08/05/2007
The protein sequence was transformed into FASTA format
Using Command Prompt and the CD, BLASTed the protein sequence. First used a vertebrate_other database (which produced only 12 significant results), before was finally able to access the nonredundant database with 57 homologous matching sequences and several different species including Homo sapiens, Danio rerio (zebra fish),Mus musculus (house mouse), Bos taurus (domesticate cow), Xenopus tropicalis (pipid frog), Rattus norvegicus (brown rat), Xenopus laevis (African clawed frog), Canis familiaris (domesticate dog), Pan troglodytes (chimpanzee), Drosophila melangaster (fruit fly), Drosophila pseudoobscura, Aedes aegypti (mosquito) and Tetraodon nigroviridis (green spotted pufferfish).
The protein seems to be conserved throughout the animal kingdom. Must have a quite important function. Hopefully when the phylogenetic tree is constructed, will get more insight into the evolution of MIF46 domain containing protein.
File that shows the significant sequences obtained from the non-redundant database.
File that shows the id numbers of the significant sequences found from the non-redundant database.
File that shows protein results
15/5/2007
Used the program ClustalX to perform a multiple alignment sequence on the 55 Id numbers (1 sequence had no Id number and 1 Id number could not be read.) When the sequences were uploaded into the ClustalX programme, performed a full alignment on the sequences. Are ready to now construct a phylogenetic tree for our protein.
Phylogenetic Tree
The first step was to perform a distance matrix calculation on the sequences. This was done by using the Phylip program on the CD. Because the program only scans the first ten characters of the Id numbers of the sequences, several sequences had to be change because they appeared to be the same. These sequences were sequences 21 & 22, 23 & 24, 32 & 33, 39 & 40 & 41 & 47, and to be on the safe side 42 & 43 need to be changed also.
22/05/2007
Original sequences and change to the sequences
21 - gi|114673074
22 - gi|114673072
21 - kept the same
22 - changed to gi|1146jpe
23 - gi|109122125
24 - gi|109122123
23 - kept the same
24 - changed to gi|1091jpe
32 - gi|123258675
33 - gi|123258677
32 - kept the same
33 - changed to gi|1232jpe
39 - gi|119609670
40 - gi|119609672
41 - gi|119609671
47 - gi|119609669
39 - kept the same
40 - changed to gi|1196jpe
41 - changed to gi|1196epj
47 - changed to gi|1196pje
42 - gi|73964908
43 - gi|73964910
42 - kept the same
43 - changed to gi|7396jpe
Redid the distance matrix calculation with the newly changed sequences. This produced a file of the calculations which was called outputsequences.phy. (I can't upload .phy files onto the page)
The first tree constructed was done using the neighbour-joining method. The output file was named neighbourjoiningspeciessequences.phy and the other output file that was produced from this method was called treealignment.ph The neighbour joining file was converted to a text file.
File that shows the neighbour joining method which produced an unrooted tree
File:Neighbourjoiningspeciessequences.txt
This data was used to convert the tree into an image by using the Phylip program.
(Still unable to upload phylogenetic tree, it is a work in progress)
When the tree was completed, bootstrapping of the tree was the next task undertaken. This was done by following the instructions on the Methods section and using the Phylogenetic tree instructions. Part A bootstrapped the sequences 100 times. Part B was producing the bootstrap distance matrices. Step C was taking this file of bootstrap values and creating a tree alingment with these values using the programme neighbor.
Finally, have been able to upload two tree diagrams that are slightly different, one has the bootstrap values
Access to Scientific Report