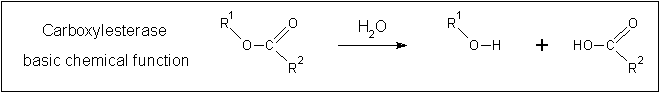Arylformamidase Discussion: Difference between revisions
Thomasparker (talk | contribs) No edit summary |
No edit summary |
||
| (12 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
Functional inference from sequence similarity was restricted due to lack of supporting literature for highest-scoring sequences. Although the function of the arylformamidase from ''Mus musculus'' has been relatively well-characterised (Pabarcus 2005), its structure has not been determined. | Functional inference from sequence similarity was restricted due to lack of supporting literature for highest-scoring sequences. Although the function of the arylformamidase from ''Mus musculus'' has been relatively well-characterised (Pabarcus 2005), its structure has not been determined. Arylformamidase proteins have not been characterised from amongst the prokaryotes, indicating it may be limited to the eukaryotes. Whilst a high-degree of conservation of the catalytic triad has been observed, there are proteins more structurally similar which provides a greater indication of function. | ||
The carboxylesterase | The carboxylesterase 2C7B was chosen for further analysis due to its high structural similarity to 2PBL. 2C7B is a thermostable carboxylesterase from a metagenomic archaeon which was isolated in a thermal environmental sample (Byun 2007). This carboxylesterase belongs to a family of bacterial hormone-sensitive lipases/esterases (HSLs). There were a number of other bacterial HSL proteins as identified by Byun, et al. (2007) found to have a high structural similarity to 2PBL. Although a relatively low sequence similarity with 2PBL was observed, regions of conserved sequence characteristic to HSL proteins aligned with 2PBL (Arpigny 1999). This may indicate that 2PBL is a member of the bacterial HSL family of lipolytic proteins. The HSL family encompasses a broad range of proteins with relatively diverse functionality which are known for their amino-acid sequence similarity to the mammalian hormone-sensitive lipase (Arpigny 1999). Contrary to this, a mammalian hormone-sensitive lipase was not returned in the BLAST search, significant evidence against 2C7B being a member of the HSL family. | ||
Further analysis revealed that 2PBL shares greater structural similarity with thermostable esterases of the HSL family compared to its other members. As discussed, thermostable proteins such as 2C7B often exist in dimers, a possiblity which remains uninvestigated for 2PBL. Both 2PBL and 2C7B have been hypothesised to exist functionally as dimers, a feature characteristic of thermostable proteins (Byun, et al. 2007). By examining the crystal structure and sequence homology, we have attempted to deduce the functional form of 2PBL. It is expected that our protein exists as a monomer indicating that it may not have thermostable properties, but this remains to be experimentally confirmed. | |||
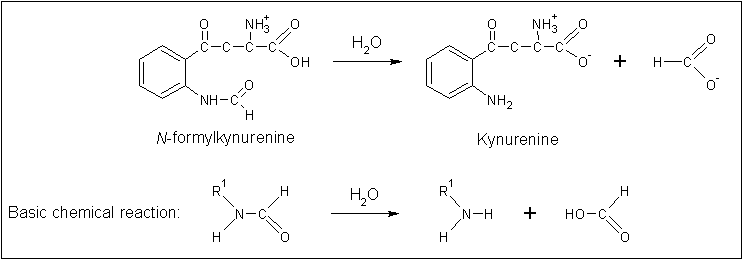
Clues to the function of 2PBL may be inferred from similarities between the fundamental reaction mechanisms of arylformamidases and carboxylesterases. Arylformamidase catalyses the hydrolysis of N-formyl-L-kynurenine to L-kynurenine, the second step in the conversion of tryptophan to nicotinic acid, NAD(H) and NADP(H) (see figure 16). Carboxylesterases catalyse the hydrolysis of a carboxylic ester into an alcohol and a carboxylic acid (see figure 17). Indeed, both arylformamidase and carboxylesterase are members of the A/B-hydrolase superfamily, suggesting that 2PBL is a A/B-hydrolase. | |||
[[Image: | [[Image:Arylformadisae_reaction.gif|centre|framed|'''Figure 16:''' ''Reaction mechanism of the mammalian arylformamidase. Produced using ChemSketch.'']] | ||
[[Image:carboxylesterase_reaction.png|centre|framed|'''Figure 17:''' ''Fundamental reaction mechanism of carboxylesterases.'']] | |||
A multiple sequence alignment revealed several conserved regions in the sequence of 2PBL across all species. This high level of conservation was noted from the Bacteria through to the Eukaryota including vertebrates, invertebrates, yeasts and moulds. Most significantly, a relatively conserved catalytic triad consisting of serine, aspartimic/glutamic acid and histidine as well as | A multiple sequence alignment revealed several conserved regions in the sequence of 2PBL across all species. This high level of conservation was noted from the Bacteria through to the Eukaryota including vertebrates, invertebrates, yeasts and moulds. Most significantly, a relatively conserved catalytic triad consisting of serine, aspartimic/glutamic acid and histidine as well as associated residues was observed in many of these sequences. The group of catalytic residues occurred in the putative functional region of 2PBL. This may be indicative of a protein scaffold used ubiquitously in the evolution of hydrolases. | ||
The phylogeny of our protein is largely consistent with traditional taxonomic groupings of organisms. Also, from the cladistics of the phylogenetic tree, there is no evidence for horizontal gene transfer (see figure ...). Thus, the delineations between prokaryotic and eukaryotic species allow us to infer that the dominant mode of inheritance of this functional region is clonal from bacteria to plantae and animalia. | |||
The A/B-hydrolase superfamily encompasses proteins with a broad range of functions. We have attempted to identify function through analysis of eukaryotic homologues, however a greater understanding of its prokaryotic function remains unclear. Identifying a specific function for 2PBL is a challenge for future research. | |||
[[Arylformamidase | Return to the main page...]] | [[Arylformamidase | Return to the main page...]] | ||
Latest revision as of 01:54, 10 June 2008
Functional inference from sequence similarity was restricted due to lack of supporting literature for highest-scoring sequences. Although the function of the arylformamidase from Mus musculus has been relatively well-characterised (Pabarcus 2005), its structure has not been determined. Arylformamidase proteins have not been characterised from amongst the prokaryotes, indicating it may be limited to the eukaryotes. Whilst a high-degree of conservation of the catalytic triad has been observed, there are proteins more structurally similar which provides a greater indication of function.
The carboxylesterase 2C7B was chosen for further analysis due to its high structural similarity to 2PBL. 2C7B is a thermostable carboxylesterase from a metagenomic archaeon which was isolated in a thermal environmental sample (Byun 2007). This carboxylesterase belongs to a family of bacterial hormone-sensitive lipases/esterases (HSLs). There were a number of other bacterial HSL proteins as identified by Byun, et al. (2007) found to have a high structural similarity to 2PBL. Although a relatively low sequence similarity with 2PBL was observed, regions of conserved sequence characteristic to HSL proteins aligned with 2PBL (Arpigny 1999). This may indicate that 2PBL is a member of the bacterial HSL family of lipolytic proteins. The HSL family encompasses a broad range of proteins with relatively diverse functionality which are known for their amino-acid sequence similarity to the mammalian hormone-sensitive lipase (Arpigny 1999). Contrary to this, a mammalian hormone-sensitive lipase was not returned in the BLAST search, significant evidence against 2C7B being a member of the HSL family.
Further analysis revealed that 2PBL shares greater structural similarity with thermostable esterases of the HSL family compared to its other members. As discussed, thermostable proteins such as 2C7B often exist in dimers, a possiblity which remains uninvestigated for 2PBL. Both 2PBL and 2C7B have been hypothesised to exist functionally as dimers, a feature characteristic of thermostable proteins (Byun, et al. 2007). By examining the crystal structure and sequence homology, we have attempted to deduce the functional form of 2PBL. It is expected that our protein exists as a monomer indicating that it may not have thermostable properties, but this remains to be experimentally confirmed.
Clues to the function of 2PBL may be inferred from similarities between the fundamental reaction mechanisms of arylformamidases and carboxylesterases. Arylformamidase catalyses the hydrolysis of N-formyl-L-kynurenine to L-kynurenine, the second step in the conversion of tryptophan to nicotinic acid, NAD(H) and NADP(H) (see figure 16). Carboxylesterases catalyse the hydrolysis of a carboxylic ester into an alcohol and a carboxylic acid (see figure 17). Indeed, both arylformamidase and carboxylesterase are members of the A/B-hydrolase superfamily, suggesting that 2PBL is a A/B-hydrolase.
A multiple sequence alignment revealed several conserved regions in the sequence of 2PBL across all species. This high level of conservation was noted from the Bacteria through to the Eukaryota including vertebrates, invertebrates, yeasts and moulds. Most significantly, a relatively conserved catalytic triad consisting of serine, aspartimic/glutamic acid and histidine as well as associated residues was observed in many of these sequences. The group of catalytic residues occurred in the putative functional region of 2PBL. This may be indicative of a protein scaffold used ubiquitously in the evolution of hydrolases.
The phylogeny of our protein is largely consistent with traditional taxonomic groupings of organisms. Also, from the cladistics of the phylogenetic tree, there is no evidence for horizontal gene transfer (see figure ...). Thus, the delineations between prokaryotic and eukaryotic species allow us to infer that the dominant mode of inheritance of this functional region is clonal from bacteria to plantae and animalia.
The A/B-hydrolase superfamily encompasses proteins with a broad range of functions. We have attempted to identify function through analysis of eukaryotic homologues, however a greater understanding of its prokaryotic function remains unclear. Identifying a specific function for 2PBL is a challenge for future research.