ATP binding domain 4 Evolution: Difference between revisions
| (49 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
== | == Evolution Analysis == | ||
== | == Methods == | ||
From | The protein that is used in this research is a putative N-type ATP pyrophosphatase from ''Pyrococcus furiosus''. This protein is predicted to be similar to ATP binding domain 4. From ‘Target Blast and Symatlas Table’, a BlastP search against the NCBI non-redundant protein database can be conducted for both sequences from ''Pyrococcus furiosus'' ATP pyrophosphatase and ''Macaca mulatta'' ATP binding domain 4. Sequences with small E value were selected and Fasta format of selected sequences were retrieved. Using these selected sequences, a multiple sequence alignment was conducted by using ClustalX. A phylogenetic tree can be constructed based on the best multiple sequence alignment so that the evolutionary relationship between species can be observed. From the phylogenetic tree, bootstrap was performed with 100 replicates. Thus, bootstrap values which indicate the branch length of the tree can be calculated. N-type ATP pyrophosphatase was also searched by using STRING database which will allow the observation of the occurrence of the protein superfamily in other species. | ||
== Results == | |||
From the BlastP search, it was found that the E value for both query sequence i.e; N-type ATP pyrophosphatase from ''Pyrococcus furiosus'' and ATP Binding Domain 4 from ''Macaca mulatta'', are very low (Figure 1 .0 and Figure 1.1). The sequences with highest E-value for ''Pyrococcus furiosus'' and the ''Macaca mulatta'' are 2e-25 and 1e-49 respectively which are very low. Therefore, all 200 sequences were aligned by using ClustalX. From the multiple sequence alignment, it was found that only one conserved residue presence in the alignment. Therefore, only extremely low E values were re-selected and unrelated sequences were ignored and deleted. Sequences were deleted in order to acheive the best multiple alignment. Based on these new re-selected sequences (88 sequences), it was found that there are seven conserved residues but only five conserved residues are significant since these residues are a part of PP-loop motif (Figure 2.0 and Figure 2.1). | |||
''' | Phylogram was constructed from Treeview software which indicates the relationship between the taxa (Figure 3.0). ''Macaca mulatta'' is closely related to ''Homo sapiens'' based on their high similarity of sequences of ATP binding domain 4. Therefore, ''Macaca mulatta'' is homologues to ''Homo sapiens''. | ||
Unrooted or radial tree was constructed from Treeview software (Figure 3.1). The tree showed the evolutionary relationship between taxa and it was found that the members of archaea and eukarya were clearly separated. Domain bacteria was absence in this tree. | |||
Bootstrap was performed to see the reliability of branching order and distance i.e: a measure of the quality of phylogenetic tree. Bootstrap value indicates the confidence level of the branching order and distance. Value which is less than 50% are meaningless. In this project, some species from the phylogenetic tree were deleted when constructing Bootstrap tree since most of the Bootstrap value are 100%. Based on bootstrap tree (Figure 4.0), most of the value are lesser than 75%. | |||
Since domain bacteria was not found in the phylogenetic tree, STRING search of N-type ATP pyrophosphatase was performed and it was found that some bacteria species actually belong to N-type ATP pyrophosphatase (Figure 5.0). However, when the sequences were aligned with that for ''Pyrococcus furiosus'' and ''Macaca mulatta'', the sequences similarity are low and the E-values are quite high. | |||
'''[[ | '''[[BlastP]]''' | ||
[[Image: | [[Image:Blastp search-1ru8.png |left|thumb|1200px|'''Figure 1.0'''. The result of BlastP search which used N-type ATP pyrophosphatase from ''Pyrococcus furiosus'' as query sequence (1RU8A)]] | ||
Figure | [[Image:Blastp-Macaca mulatta.png |left|thumb|1200px|'''Figure 1.1'''. The result of BlastP search which used protein sequence that is predicted to be similar to ATP binding domain 4 from ''Macaca mulatta'']] <BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR> | ||
'''[[Multiple sequence alignment-ClustalX]]''' | |||
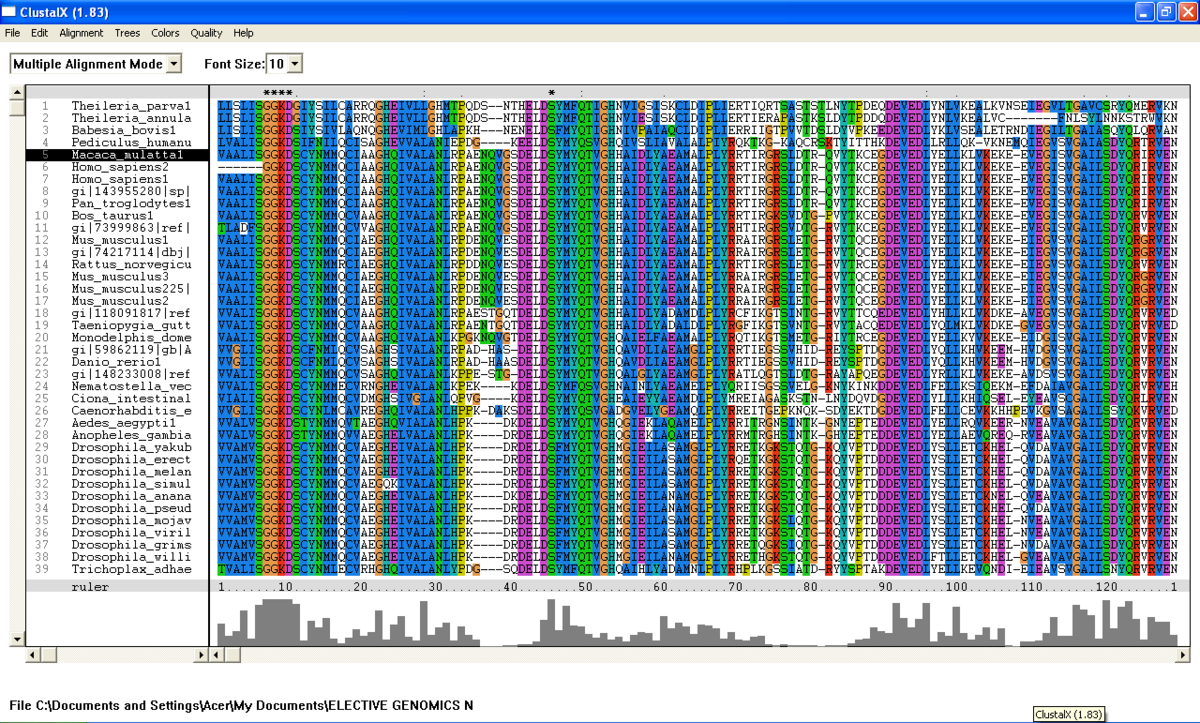
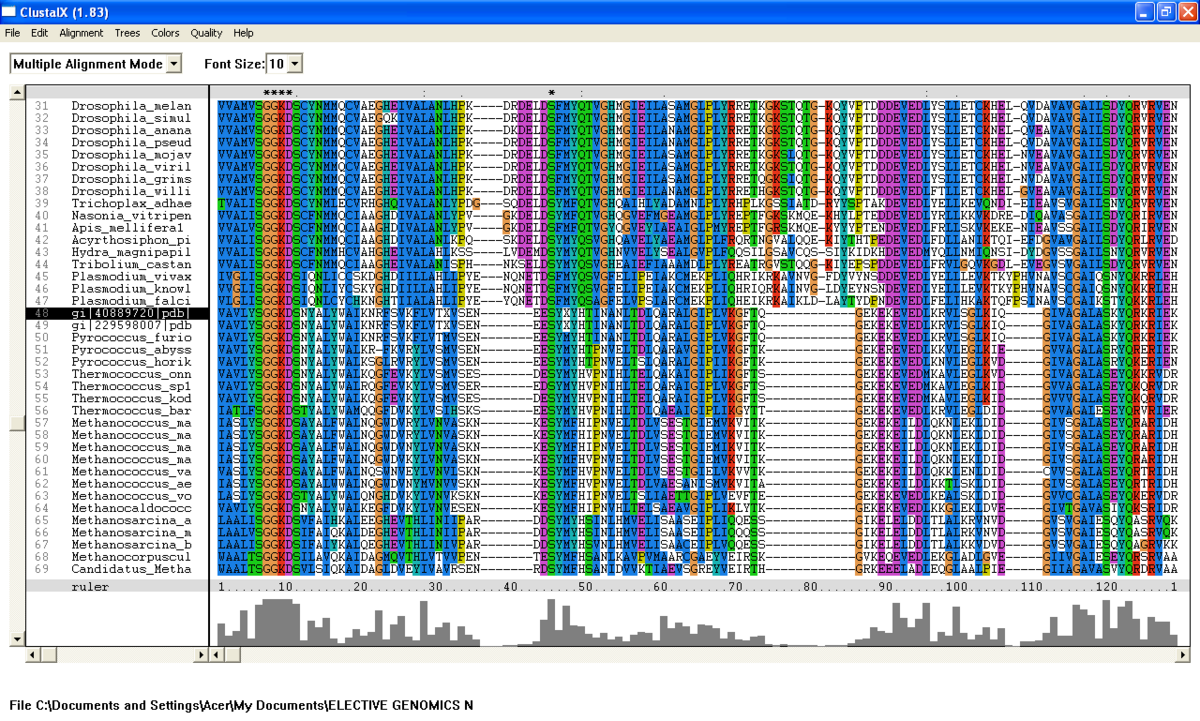
[[Image:Clustalx taken from residue 98-350.png |left|thumb|1200px|'''Figure 2.0'''. Multiple sequence alignment obtained from ClustalX. Section of msa revealed five important residue which are conserved across species in archaea and eukarya.]] | |||
Figure | [[Image:Clustalx taken from residue 98-350 Part2.png |left|thumb|1200px|'''Figure 2.1'''. Multiple sequence alignment obtained from ClustalX. Section of msa revealed five important residue which are conserved across species in archaea and eukarya. gi:40889720 denotes N-type ATP pyrophosphatase in ''Pyrococcus furiosus.'' ]] | ||
'''[[ | '''[[Phylogenetic tree]]''' | ||
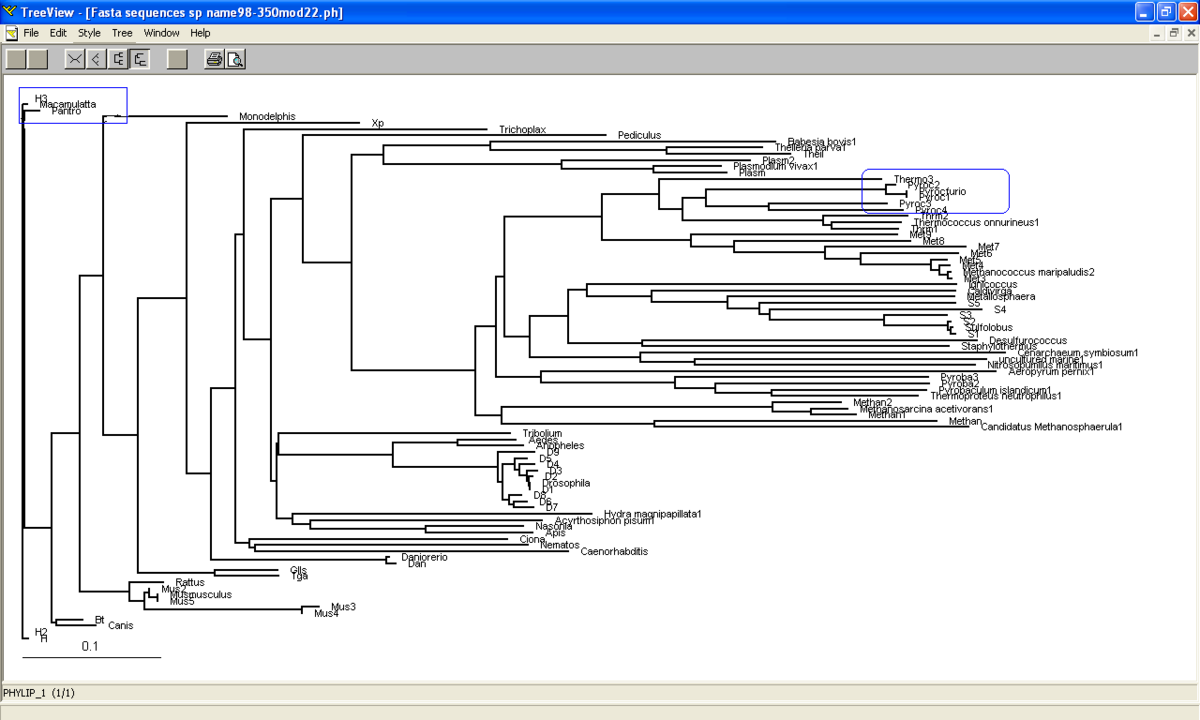
[[Image:Phylogram.png]] | [[Image:Phylogram.png |left|thumb|1200px|'''Figure 3.0'''. Phylogram constructed from Treeview.]] | ||
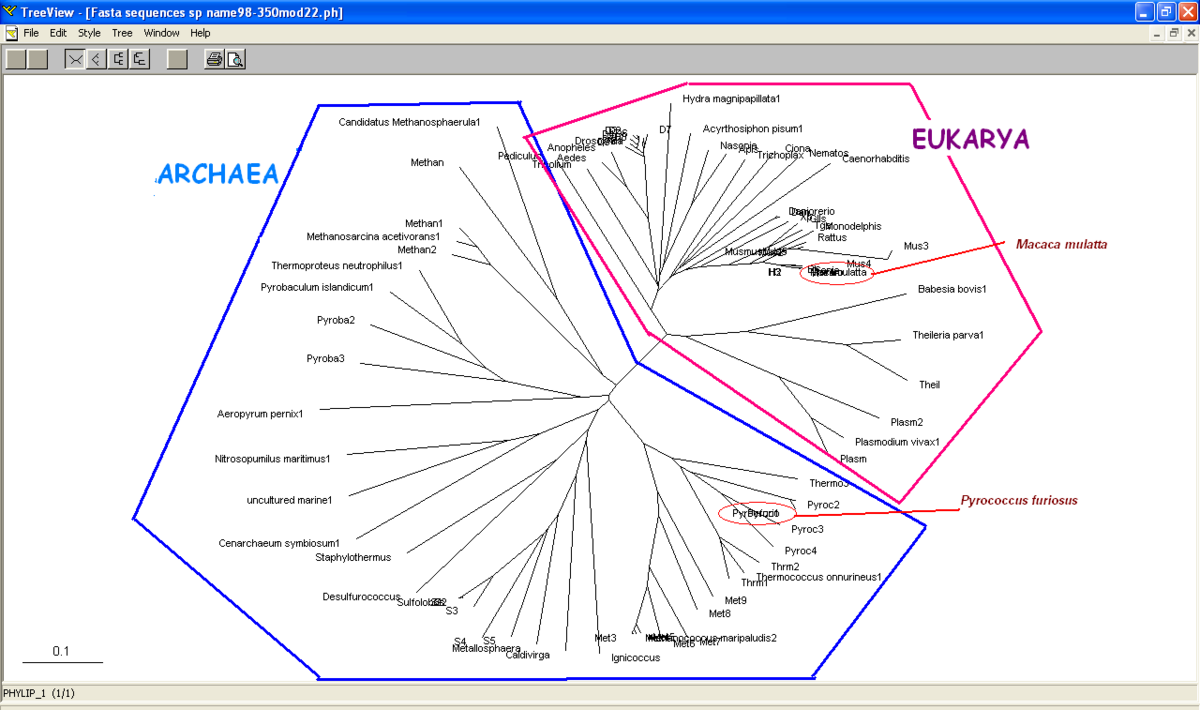
[[Image:Unrooted tree ATP Binding Domain 4.PNG]] | [[Image:Unrooted tree ATP Binding Domain 4.PNG |left|thumb|1200px|'''Figure 3.1'''. Unrooted phylogenetic tree of species with low E-value from a blastP search of N-type ATP pyrophosphatase from ''Pyrococcus furiosus'' and ATP Binding Domain 4 from ''Macaca mulatta''. Members of Eukarya and Archaea are clearly separated and clustered together. No members of Bacteria present in this phylogenetic tree.]] | ||
'''[[Bootstrap]]''' | '''[[Bootstrap]]''' | ||
[[Image:Boostrap.png | [[Image:Boostrap.png |left|thumb|1200px| '''Figure 4.0'''. Phylogenetic tree with Bootstrap values. The tree was constructed by using Mega4. The following are gi number and their respected species and protein; gi:143955280 = ATP-binding domain containing protein 4 from ''Homo sapiens'', gi: 73999863 = protein predicted to be similar to CG1578-PA from ''Canis familiaris'', gi:7421711 = unnamed protein product from ''Mus musculus'', gi: 1180918 = protein predicted to be similar to MGC83562 protein from ''Gallus gallus'', gi:59862119 = Zgc:110758 protein from ''Danio rerio'', gi:4088972 = putative N-type ATP pyrophosphatase and gi: 2295980 = Chain A, crystal structure of an N-type ATP pyrophosphatase in complex with Amp from ''Pyrococcus furiosus''.]]<BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR> | ||
<BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR> | |||
Figure | <BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR> | ||
'''[[STRING]]''' | '''[[STRING]]''' | ||
[[Image:String_ATP_pyrophosphatase.png]] | [[Image:String_ATP_pyrophosphatase.png |left|thumb|1200px|'''Figure 5.0''': Result from STRING database illustrates that some Bacteria species still have protein which belong to N-type ATP pyrophosphatase superfamily.]] | ||
== Discussion == | |||
'''Multiple sequence alignment''' | '''Multiple sequence alignment''' | ||
From the multiple sequence alignment, there are seven conserved regions found in all species from Domain Eukarya and Archaea (based on | From the multiple sequence alignment, there are seven conserved regions found in all species from Domain Eukarya and Archaea (based on BlastP search) but only five conserved residues which are significant to the function and structure of the protein since these residues are identified to be conserved amino acid sequence motif for P-loop of nucleotide binding domains which might be important in phosphate binding (Bork and Koonin, 1994). These conserved residues are Serine-12, Glysine-13 and Glysine-14, Lysine-15 and Aspartic acid-16 . Since this motif presence in uncharacterized ATP pyrophosphatase domain, the motif is called PP motif and can be written as S-G(2)-K-D-[GS]. PP-loop motif is a modified version of the P-loop of nucleotide binding domain that is involved in phosphate binding (Bork and Koonin,1994). However, based on the multiple sequence alignment, the amino acids for PP-loop that are conserved across all species in Eukarya and Archaea are Glysine-12 and Glysine-14. Substitution of Serine with Threonine in ''Pyrobaculum arsenaticum, Pyrobaculum calidifontis, Thermoproteus neutrophilis, Pyrobaculum islandicum'' and ''Staphlothermus marinus'', are conserved since both amino acids are polar uncharged amino acid, suggesting that this residue is also important. However, this residue is not conserved in ATP Binding Domain 4 in ''Homo sapiens'' which is only encoded by 259 amino acid sequences and the sequence started from Glysine-13. Moreover, amino acid in residue 17 is also one of the important residues in PP motif. This residue is conserved across all species except for ''Theileria parva'' and ''Theileria annulata'' where Serine is substituted with Glysine. | ||
BlastP search revealed the very low E value from many sequences from different species. These sequences appear to be encoding different group of proteins such as ATP pyrophosphatase, ATP-binding protein, ATPase and endoribonuclease. Although these proteins are responsible for different functions, the similarities of sequence between the proteins across species are very high (as indicated by E value) and the sequences which are conserved are a part of PP motif, suggesting that these proteins descend from a common ancestral sequence and therefore are paralogs. Such phenomenon could be as the result of duplication of genes within a genome. | |||
'''Phylogenetic tree and Bootstrap''' | |||
''' | The phylogenetic tree and bootstrap revealed that PP-loop motif from ATP binding domain 4 (belongs to N-type ATP pyrophosphatase/ATPases) and other related proteins are found in species of Archaea and Eukarya suggesting that this motif is highly conserved throughout evolution. However, Bacteria found to be lacking of this conserved sequences since none of the species belongs to Domain Bacteria. Nevertheless, based on STRING: functional protein association networks, some bacteria species still have protein sequences which belong to N-type ATP pyrophosphatase superfamily. BlastP was conducted to compare the protein sequences of ATPases from ''Pyrococcus furiosus'' and the bacteria species which under the clan of N-type ATP pyrophosphatase. The E value obtained from the result are below the E-value that is used as the cut-off-score. The bacteria species are ''Fusobacteruim nucleatum'' (1e-22), ''Caldicellulosiruptor saccharolyticus'' (1e-13),''Campylobacter jejuni'' (4e-19),''Chromobacterium violaceum'' (1e-21)and ''Polynucleobacter sp.'' (1e-24). These E value fall below the lowest E value based on BlastP from ''Pyrococcus furiosus'' and ''Macacca mulatta'' i.e: 5e-50 and 2e-25 respectively. These bacteria species can not be found in BlastP on the first place because the E value are quite high which indicate the sequence similarity between bacteria species and ''Pyrococcus furiosus'' and ''Macaca mulatta'' are very low. These suggested that although some bacteria species still have protein sequences which belong to N-type ATP pyrophosphtase superfamily, they are distantly related to both archaea and eukarya. Based on the high protein sequence similarity between Archaea and Eukarya, it can be suggested that Archaea and Eukarya are closely related compared to that for Bacteria. | ||
Based on unrooted tree, the relatedness of the taxa and their relationship can be illustrated but the last common ancestor and how the species evolved cannot be observed. It was found that the taxa are clearly grouped into Domain Archaea and Eukarya. Moreover, in Eukarya, the taxa are nicely clustered according to parasites, vertebrates and invertebrates. Domain Bacteria is missing in this tree suggesting that Bacteria are distantly related to Archaea and Eukarya based on the sequence of ATP Binding Domain 4. | |||
Phylogram (figure 3.0) is another way to illustrate the evolution of the taxa where the relationships between the taxa and the time or rate of evolution can be observed. | |||
Bootstrap was conducted to test the reliability of the branching order of the phylogenetic tree. Based on the bootstrap value, we can be confident with the order of the phylogenetic tree thus allowing us to determine where speciation events occur. Bootstrap worked by performing 'pseudoreplicates' of multiple sequence alignments and in this project, 100 replicates was performed. The bootstrap tree can then be generated thus comparing the branching orders and distances of phylogenetic tree. The bootstrap value for each branch are obtained in percentage which indicate the confidence of the branch being correct. If the value of bootstrap is less than 75%, the branching order is not very reliable and meaningless. If the value is between 90% and above, we can be confident that the branching orders are correct. Based on the bootstrap result, it was found that most of the value are lower than 75% suggesting that the branching patterns and distances are not reliable. Therefore, another phylogenetic tree need to be built in order to increase the reliability of the branching pattern and distances of the tree. | |||
Based on unrooted tree | Based on unrooted tree, it was found members of Domain Eukarya are nicely cluster at one side without any presence of other species from different domain i.e: members from different domains are well-seperated. Therefore, it can be suggested that the evolutionary model for the PP-loop motif is hold since there is no evidence of the occurrence of lateral gene transfer. | ||
These result support theory for origins of eukaryotes which suggested chimeric features of eukaryote genome (fusion of archaea and bacteria). Study which involved whole-genome-sequence data to test this theory, indicates Eukaryote genome is a chimera of genes most similar to that in Archaea and Bacteria. This study used 'homology-hit' analysis in which the genes from eukaryotes from different classes matching to nearest homology genes in Archaea and Bacteria. It was found that informational genes are closely related to Archaea whereas operational genes are closely related to bacteria (Horiike et al., 2001). Moreover, study conducted by Brown and Doolittle which involved analysing geneolgies from 66 protein-coding genes from members of all three domains of life found that Arginosuccinate synthase from Eukaryotes are closely related to Archaea (analysis on structural similarity found that the conserved region of Arginosuccinate synthase are similar to that of ATP pyrophosphatase)(Katz, 1998). Hence, finding from previous studies support the evolutionary relationship between the three Domain of life based on ATP Binding Domain 4 which suggested that that Archaea and Eukarya are closely related. | |||
[[Image:Horiike.PNG|left|thumb|1200px|'''Figure 6.0''': Chimeric nature of eukarya based on geneologies. Figure adapted from Katz, 1998 ]] | |||
<BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR> | |||
| Line 87: | Line 88: | ||
[[ATP binding domain 4 Evolution | Evolutionary Analysis]]|<BR> | [[ATP binding domain 4 Evolution | Evolutionary Analysis]]|<BR> | ||
[[ATP binding domain 4 Discussions | Discussions]]| | [[ATP binding domain 4 Discussions | Discussions]]| | ||
[[ATP binding domain 4 Conclusion | Conclusion]] | | |||
[[ATP binding domain 4 References | References]] | [[ATP binding domain 4 References | References]] | ||
[[ATP binding domain 4 | Back to Main ATP binding domain 4 pages]] | [[ATP binding domain 4 | Back to Main ATP binding domain 4 pages]] | ||
Latest revision as of 09:01, 14 June 2009
Evolution Analysis
Methods
The protein that is used in this research is a putative N-type ATP pyrophosphatase from Pyrococcus furiosus. This protein is predicted to be similar to ATP binding domain 4. From ‘Target Blast and Symatlas Table’, a BlastP search against the NCBI non-redundant protein database can be conducted for both sequences from Pyrococcus furiosus ATP pyrophosphatase and Macaca mulatta ATP binding domain 4. Sequences with small E value were selected and Fasta format of selected sequences were retrieved. Using these selected sequences, a multiple sequence alignment was conducted by using ClustalX. A phylogenetic tree can be constructed based on the best multiple sequence alignment so that the evolutionary relationship between species can be observed. From the phylogenetic tree, bootstrap was performed with 100 replicates. Thus, bootstrap values which indicate the branch length of the tree can be calculated. N-type ATP pyrophosphatase was also searched by using STRING database which will allow the observation of the occurrence of the protein superfamily in other species.
Results
From the BlastP search, it was found that the E value for both query sequence i.e; N-type ATP pyrophosphatase from Pyrococcus furiosus and ATP Binding Domain 4 from Macaca mulatta, are very low (Figure 1 .0 and Figure 1.1). The sequences with highest E-value for Pyrococcus furiosus and the Macaca mulatta are 2e-25 and 1e-49 respectively which are very low. Therefore, all 200 sequences were aligned by using ClustalX. From the multiple sequence alignment, it was found that only one conserved residue presence in the alignment. Therefore, only extremely low E values were re-selected and unrelated sequences were ignored and deleted. Sequences were deleted in order to acheive the best multiple alignment. Based on these new re-selected sequences (88 sequences), it was found that there are seven conserved residues but only five conserved residues are significant since these residues are a part of PP-loop motif (Figure 2.0 and Figure 2.1).
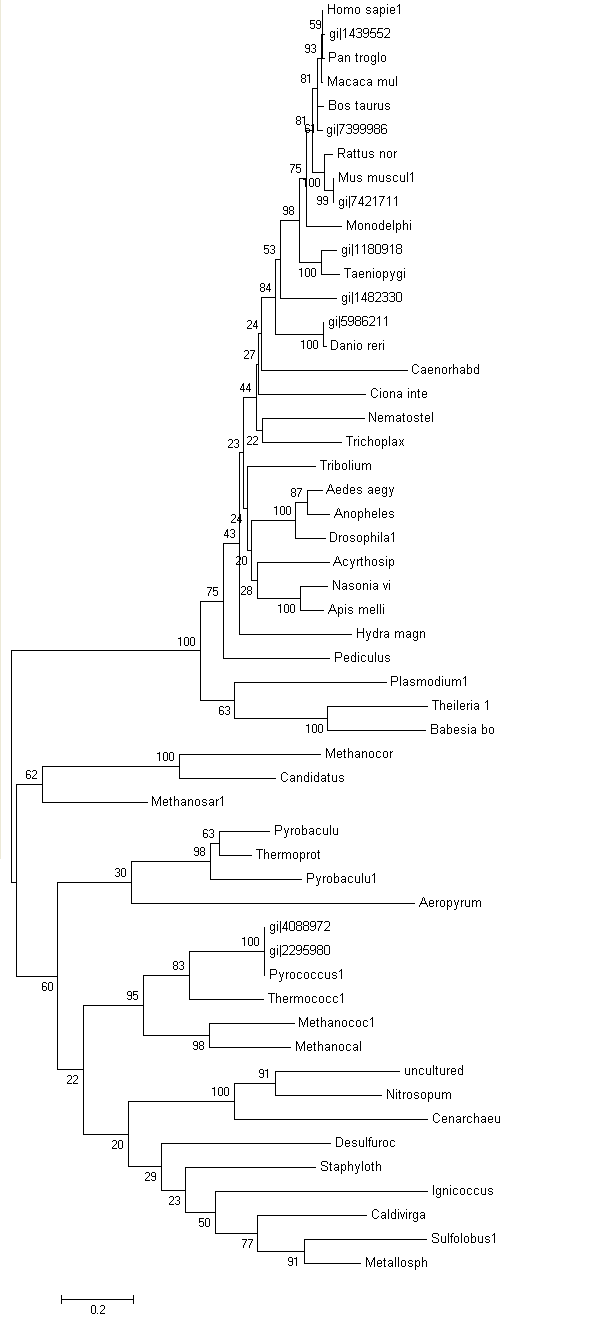
Phylogram was constructed from Treeview software which indicates the relationship between the taxa (Figure 3.0). Macaca mulatta is closely related to Homo sapiens based on their high similarity of sequences of ATP binding domain 4. Therefore, Macaca mulatta is homologues to Homo sapiens.
Unrooted or radial tree was constructed from Treeview software (Figure 3.1). The tree showed the evolutionary relationship between taxa and it was found that the members of archaea and eukarya were clearly separated. Domain bacteria was absence in this tree.
Bootstrap was performed to see the reliability of branching order and distance i.e: a measure of the quality of phylogenetic tree. Bootstrap value indicates the confidence level of the branching order and distance. Value which is less than 50% are meaningless. In this project, some species from the phylogenetic tree were deleted when constructing Bootstrap tree since most of the Bootstrap value are 100%. Based on bootstrap tree (Figure 4.0), most of the value are lesser than 75%.
Since domain bacteria was not found in the phylogenetic tree, STRING search of N-type ATP pyrophosphatase was performed and it was found that some bacteria species actually belong to N-type ATP pyrophosphatase (Figure 5.0). However, when the sequences were aligned with that for Pyrococcus furiosus and Macaca mulatta, the sequences similarity are low and the E-values are quite high.
Multiple sequence alignment-ClustalX
Discussion
Multiple sequence alignment
From the multiple sequence alignment, there are seven conserved regions found in all species from Domain Eukarya and Archaea (based on BlastP search) but only five conserved residues which are significant to the function and structure of the protein since these residues are identified to be conserved amino acid sequence motif for P-loop of nucleotide binding domains which might be important in phosphate binding (Bork and Koonin, 1994). These conserved residues are Serine-12, Glysine-13 and Glysine-14, Lysine-15 and Aspartic acid-16 . Since this motif presence in uncharacterized ATP pyrophosphatase domain, the motif is called PP motif and can be written as S-G(2)-K-D-[GS]. PP-loop motif is a modified version of the P-loop of nucleotide binding domain that is involved in phosphate binding (Bork and Koonin,1994). However, based on the multiple sequence alignment, the amino acids for PP-loop that are conserved across all species in Eukarya and Archaea are Glysine-12 and Glysine-14. Substitution of Serine with Threonine in Pyrobaculum arsenaticum, Pyrobaculum calidifontis, Thermoproteus neutrophilis, Pyrobaculum islandicum and Staphlothermus marinus, are conserved since both amino acids are polar uncharged amino acid, suggesting that this residue is also important. However, this residue is not conserved in ATP Binding Domain 4 in Homo sapiens which is only encoded by 259 amino acid sequences and the sequence started from Glysine-13. Moreover, amino acid in residue 17 is also one of the important residues in PP motif. This residue is conserved across all species except for Theileria parva and Theileria annulata where Serine is substituted with Glysine.
BlastP search revealed the very low E value from many sequences from different species. These sequences appear to be encoding different group of proteins such as ATP pyrophosphatase, ATP-binding protein, ATPase and endoribonuclease. Although these proteins are responsible for different functions, the similarities of sequence between the proteins across species are very high (as indicated by E value) and the sequences which are conserved are a part of PP motif, suggesting that these proteins descend from a common ancestral sequence and therefore are paralogs. Such phenomenon could be as the result of duplication of genes within a genome.
Phylogenetic tree and Bootstrap
The phylogenetic tree and bootstrap revealed that PP-loop motif from ATP binding domain 4 (belongs to N-type ATP pyrophosphatase/ATPases) and other related proteins are found in species of Archaea and Eukarya suggesting that this motif is highly conserved throughout evolution. However, Bacteria found to be lacking of this conserved sequences since none of the species belongs to Domain Bacteria. Nevertheless, based on STRING: functional protein association networks, some bacteria species still have protein sequences which belong to N-type ATP pyrophosphatase superfamily. BlastP was conducted to compare the protein sequences of ATPases from Pyrococcus furiosus and the bacteria species which under the clan of N-type ATP pyrophosphatase. The E value obtained from the result are below the E-value that is used as the cut-off-score. The bacteria species are Fusobacteruim nucleatum (1e-22), Caldicellulosiruptor saccharolyticus (1e-13),Campylobacter jejuni (4e-19),Chromobacterium violaceum (1e-21)and Polynucleobacter sp. (1e-24). These E value fall below the lowest E value based on BlastP from Pyrococcus furiosus and Macacca mulatta i.e: 5e-50 and 2e-25 respectively. These bacteria species can not be found in BlastP on the first place because the E value are quite high which indicate the sequence similarity between bacteria species and Pyrococcus furiosus and Macaca mulatta are very low. These suggested that although some bacteria species still have protein sequences which belong to N-type ATP pyrophosphtase superfamily, they are distantly related to both archaea and eukarya. Based on the high protein sequence similarity between Archaea and Eukarya, it can be suggested that Archaea and Eukarya are closely related compared to that for Bacteria.
Based on unrooted tree, the relatedness of the taxa and their relationship can be illustrated but the last common ancestor and how the species evolved cannot be observed. It was found that the taxa are clearly grouped into Domain Archaea and Eukarya. Moreover, in Eukarya, the taxa are nicely clustered according to parasites, vertebrates and invertebrates. Domain Bacteria is missing in this tree suggesting that Bacteria are distantly related to Archaea and Eukarya based on the sequence of ATP Binding Domain 4. Phylogram (figure 3.0) is another way to illustrate the evolution of the taxa where the relationships between the taxa and the time or rate of evolution can be observed.
Bootstrap was conducted to test the reliability of the branching order of the phylogenetic tree. Based on the bootstrap value, we can be confident with the order of the phylogenetic tree thus allowing us to determine where speciation events occur. Bootstrap worked by performing 'pseudoreplicates' of multiple sequence alignments and in this project, 100 replicates was performed. The bootstrap tree can then be generated thus comparing the branching orders and distances of phylogenetic tree. The bootstrap value for each branch are obtained in percentage which indicate the confidence of the branch being correct. If the value of bootstrap is less than 75%, the branching order is not very reliable and meaningless. If the value is between 90% and above, we can be confident that the branching orders are correct. Based on the bootstrap result, it was found that most of the value are lower than 75% suggesting that the branching patterns and distances are not reliable. Therefore, another phylogenetic tree need to be built in order to increase the reliability of the branching pattern and distances of the tree.
Based on unrooted tree, it was found members of Domain Eukarya are nicely cluster at one side without any presence of other species from different domain i.e: members from different domains are well-seperated. Therefore, it can be suggested that the evolutionary model for the PP-loop motif is hold since there is no evidence of the occurrence of lateral gene transfer.
These result support theory for origins of eukaryotes which suggested chimeric features of eukaryote genome (fusion of archaea and bacteria). Study which involved whole-genome-sequence data to test this theory, indicates Eukaryote genome is a chimera of genes most similar to that in Archaea and Bacteria. This study used 'homology-hit' analysis in which the genes from eukaryotes from different classes matching to nearest homology genes in Archaea and Bacteria. It was found that informational genes are closely related to Archaea whereas operational genes are closely related to bacteria (Horiike et al., 2001). Moreover, study conducted by Brown and Doolittle which involved analysing geneolgies from 66 protein-coding genes from members of all three domains of life found that Arginosuccinate synthase from Eukaryotes are closely related to Archaea (analysis on structural similarity found that the conserved region of Arginosuccinate synthase are similar to that of ATP pyrophosphatase)(Katz, 1998). Hence, finding from previous studies support the evolutionary relationship between the three Domain of life based on ATP Binding Domain 4 which suggested that that Archaea and Eukarya are closely related.
Abstract|
Introductions|
Methods|
Structural Analysis|
Functional Analysis|
Evolutionary Analysis|
Discussions|
Conclusion |
References