COASY results: Difference between revisions
No edit summary |
Rachaelslade (talk | contribs) |
||
| (184 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
==Structure of Coenzyme A Synthase== | ==Structure of Coenzyme A Synthase== | ||
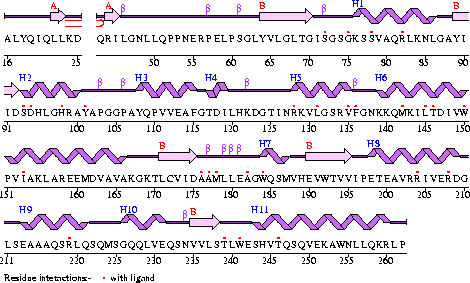
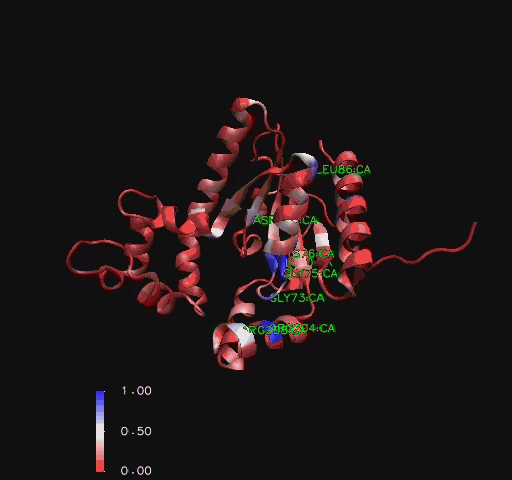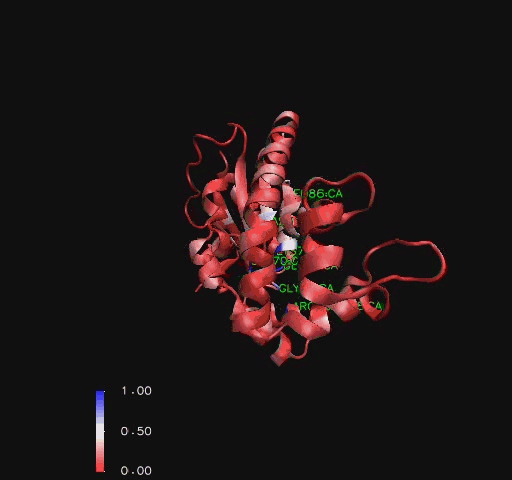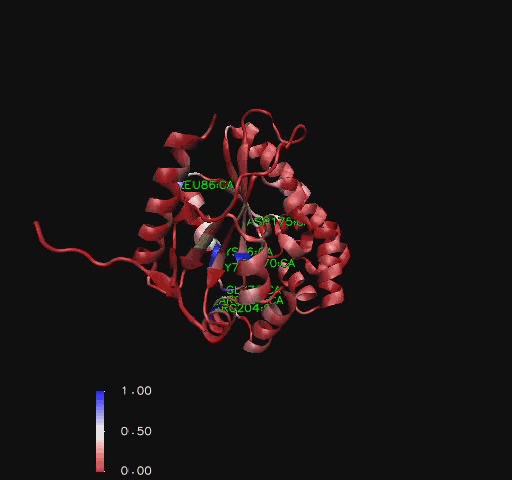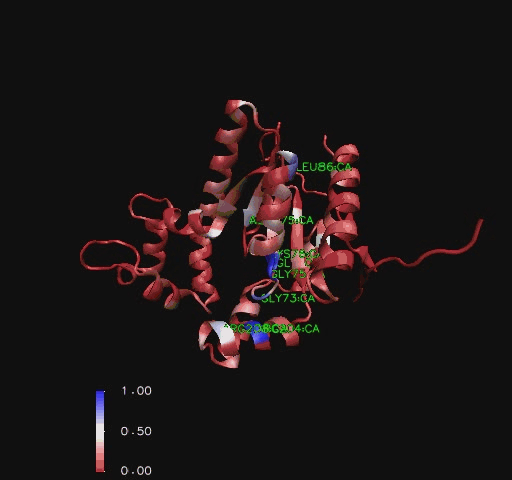
Conezyme A Synthase is structurally composed of seven strands, eleven helices and thirteen beta turns (EMBL EBI, 2005) (see [[COASY_results# | Conezyme A Synthase is structurally composed of seven strands, eleven helices and thirteen beta turns (EMBL EBI, 2005) (see [[COASY_results#Structure of Coenzyme A Synthase|Figure 2]]). Analysis of structurally related proteins (Holm & Sander, 1993) showed a trend for transferase (RCSB, 2007) class proteins with Rossmann class folds (Rossmann, 1973). The Rossmann topologies of folds are an alpha-beta class fold that forms a three or more layer beta strand sandwich alternating with alpha helices (beta-alpha-beta-alpha-beta). PFAM classification placed these into either the Cytidylyltransferase or Dephospho-CoA kinase familes, matching the two domains of Coenzyme A Synthase. Those classified under the Dephospho-CoA kinase type were commonly of the P-loop containing nucleotide triphosphate hydrolases (Sanger Institute, 2005) homology whilst those classified as Cytidylyltransferase were of the Tyrosol-Transfer RNA Synthetase (Sanger Institute, 2005). As the majority of the structurally related proteins identified contained the DPCK domain solely, and the motif for a P-loop ([[COASY_results#Functional Sites Found By Pattern Search|Table 1]]) was identified in the Conzyme A Synthase sequence (Bairoch, Bucher, & Hofmann, 1997) it is suggested that Coenzyme A Synthase is also of the P-loop containing nucleotide triphosphate hydrolases homology of folds. | ||
<BR> | <BR> | ||
{| | |||
|+ PROSITE Motif search (Bairoch, Bucher, & Hofmann, 1997) results on Coenzyme A Synthase: Mus | |||
[[Image:COASYSecondary structure with bind sites.gif|framed|'''Figure 2'''<BR>The secondary structure of ''Mus musculus'' with indicated ligand interaction sites (EMBL EBI, 2005).|none]]<BR> | |||
==Localisation Expression of Coenzyme A Synthase== | |||
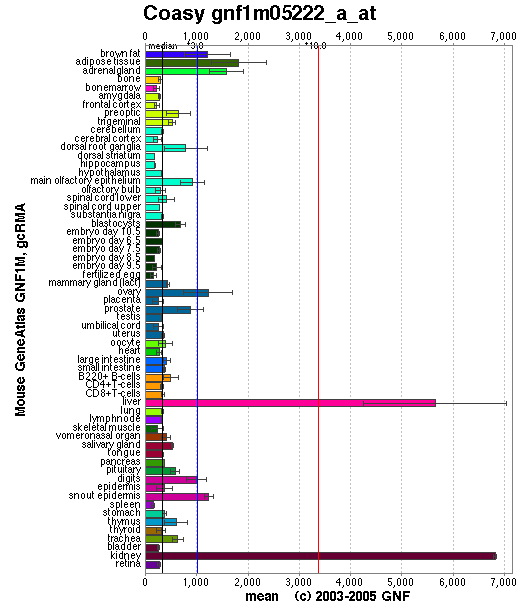
Baseline expression of CoAsy was present in all tissues. Expression was slightly elevated in adipose tissue, and highly elevated in the liver and kidney (See [[COASY_results#Localisation Expression of Coenzyme A Synthase|Figure 3]]). Baseline expression of the other enzymes involved in the CoAsy pathway was also present in all tissues. However, expression of these enzymes was otherwise unlike that of CoA, as they were not particularly elevated in the liver and kidney (data not shown). | |||
[[Image:CoA synthase localisation.png|framed|'''Figure 3 '''<BR> CoA Synthase Expression. Reproduced from Genomics Institute of Novartis Research Foundation (2007).|none]]<BR> | |||
==Domain and Structural Analysis== | |||
Local sequence alignment showed that CoAsy Chain A residues 13-281 corresponded to residues 295-563 on full length CoAsy. This indicates that a 295 amino acid stretch extending from residues 1 to 295 on the full length protein was cut out during structural analysis and was not present on chain A. | |||
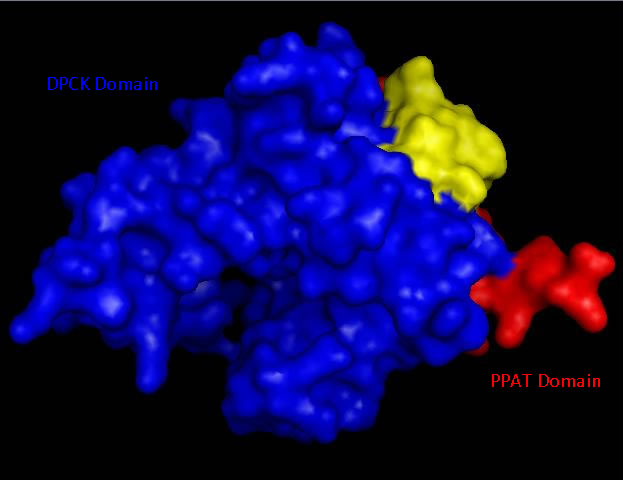
Bioinformatic analysis using NCBI Entrez protein indicated that mouse CoAsy (full length) possess two domains, a PPAT and a DPCK domain. The PPAT domain extended from residues 194-338, and the DPCK domain from residues 359-536. PFAM analysis of CoAsy chain A also indicated the presence of two domains, the DPCK domain from residues 64-242 and a CTP transferase 2 domain (part of the cytidylyltransferase family, of which PPAT is a member) extending from residues 1-44, which corresponds to a fragment of the PPAT domain. The position of these domains on CoAsy chain A is shown in [[COASY_results#Domain and Structural Analysis|Figure 4]]. Profunc analysis of CoAsy chain A detected the DPCK domain, but not the fragment of the PPAT domain. | |||
[[Image:Domains_2F6r.jpg|framed|'''Figure 4 '''<BR> Surface structure of ''Mus musculus'' Coenzyme A Synthase with Phosphopantetheine adenylyltransferase (PPAT) and Dephospho-CoA kinase (DPCK) domains marked in red and blue respectfully.|none]]<BR> | |||
==Structural Elements and Functional Binding Sites of Coenzyme A Synthase== | |||
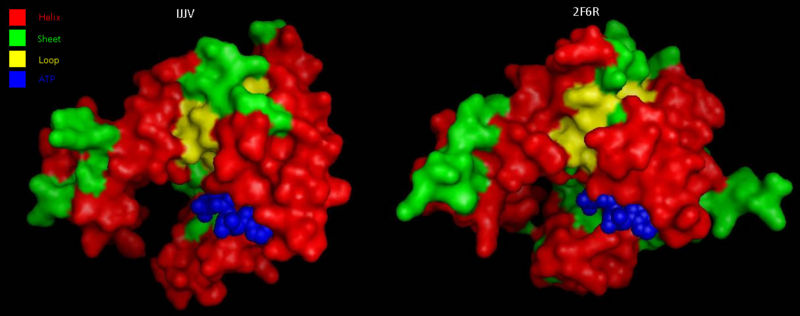
Comparison to CoAsy’s closest structurally related protein, Dephospho-CoenzymeA Kinase, finds that whilst it shares only twenty percent of its sequence, it is structurally very similar ([[COASY_results#Structural Elements and Functional Binding Sites of Coenzyme A Synthase|Figure 5]]). Apart from the lack of a PPAT domain on the Dephospho-CoenzymeA Kinase molecule (see [[COASY_results#Domain and Structural Analysis|Figure 4]] for comparative location of PPAT), all structural features are roughly the same between the two proteins. As helices, sheets and loops stay relatively constant between the two, and Dephospho-CoenzymeA Kinase has a related function to CoAsy it can be assumed that the location of the ATP binding site on the DPCK domain of CoAsy is the same as that on Dephospho-CoenzymeA Kinase. | |||
[[Image:COASYSecondary structure comparsion.jpg|thumb|800px|'''Figure 5 '''<BR> Structural comparison of Dephospho-COA Kinase (IJJV) and Bifunctional coenzyme A synthase from ''Mus musculus'' (2F6R) with bound ATP.|none]]<BR> | |||
===Electrostatic Surface Potential=== | |||
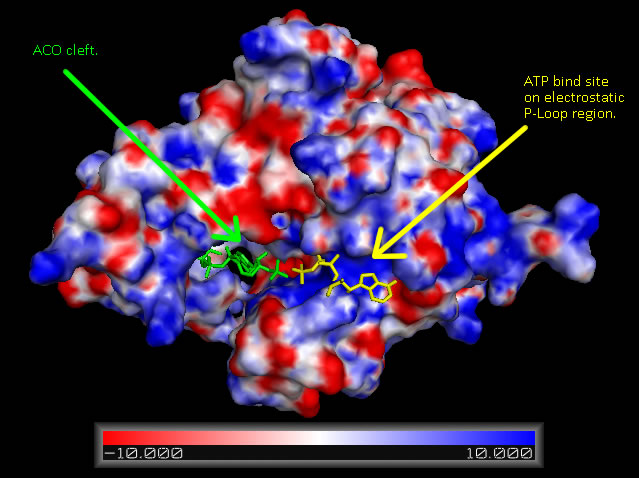
Analysis of electrostatic surface potential maps for CoAsy finds that there is a strongly positive charged region, between residues 70 and 80, on the first helix that lines up with the proposed ATP binding site ([[COASY_results#Electrostatic Surface Potential|Figure 6]]). It can also be noted that the inner wall of the major cleft alternates strongly between a positive and negative charge and this lines up with the ACO ligand that is included in the 2F6R PDB entry at this location (EMBL EBI, 2005).<BR> | |||
[[Image:COASYApbs_mapped_binding_cradle_shown_with_aligned_DPCK_IJJV_and_associated_ATP_ligand_biniding.jpg|framed|'''Figure 6 '''<BR> Surface map of ''Mus musculus'' Coenzyme A Synthase with electrostatic charge indicated, ATP ligand is included at P-Loop bind site of DPCK domain and ACO at the proposed Dephospho Coenzyme A Kinase cleft.|none]]<BR> | |||
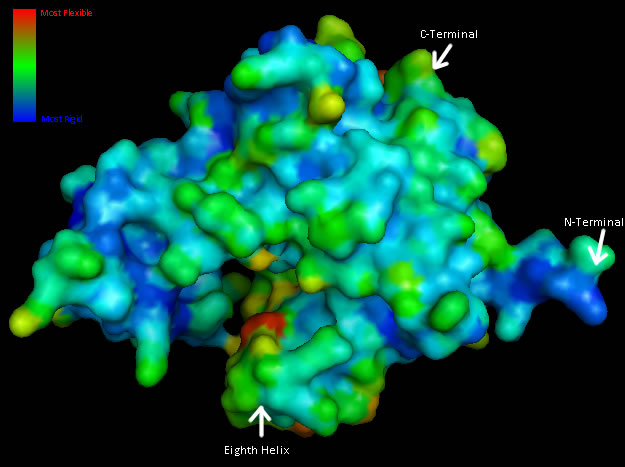
===Residue Flexibility=== | |||
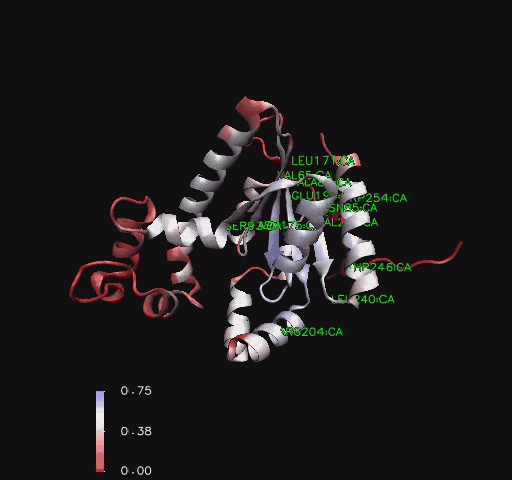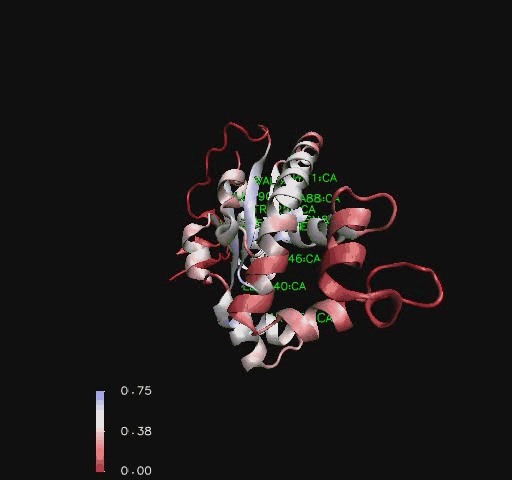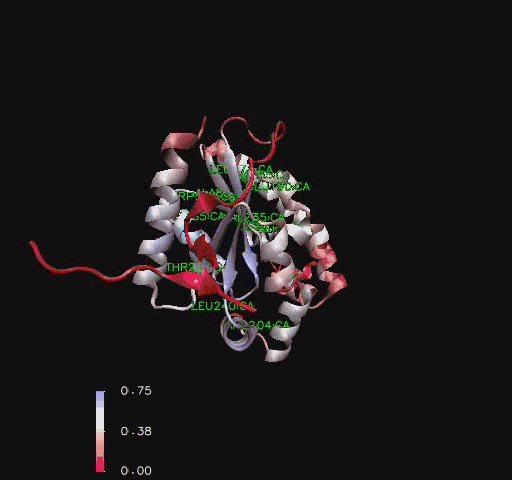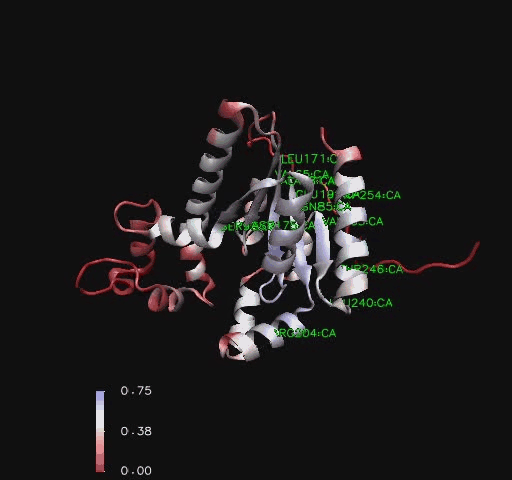
Structural analysis of Coenzyme A Synthase B-Factor scores finds it to be most stable close to the center, and on the N-terminal of the protein. The most flexible regions are on the eighth helix and on the outer edges of the protein ([[COASY_results#Residue Flexibility|Figure 7]]). The position of most flexibility on the protein is at the top of the eight loop, on the DPCK domain, and this lies between both the bind site for ATP and the location of ACO on the 2F6R PDB entry (EMBL EBI, 2005).<BR> | |||
[[Image:COASYBfactor.jpg|framed|'''Figure 7 '''<BR> Coenzyme A Synthase ''Mus musculus'' surface structure marked by B-Factor.|none]]<BR> | |||
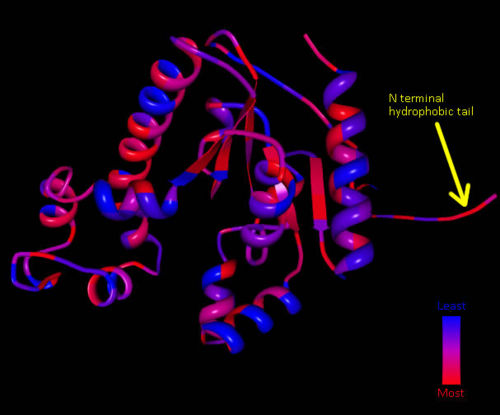
===Hydrophobicity=== | |||
Hydrophobic mapping of ''Mus musculus'' Coenzmye A Synthase finds a number of hydrophobic stretches including the inner strands and the N terminal [[COASY_results#Hydrophobicity|Figure 8]]. A study by (Zhyvoloup, et al., 2003) also found that the ''Mus musculus'' CoAsy had a hydrophobic N terminal and that this is a conserved feature for CoAsy across a broad number of species [[COASY_results#Hydrophobicity|Figure 9]].<BR> | |||
[[Image:COASYHydro2.jpg|thumb|500px|'''Figure 8 '''<BR> Coenzyme A Synthase ''Mus musculus'' ribbon structure showing hydrophobic regions.|none]]<BR> | |||
[[Image:COASYHydro.jpg||thumb|500px|'''Figure 9 '''<BR> Sequence alignment of N domain (first 30 residues) of Coenzyme A Synthase proteins (bottom) with hydrophobic residues shaded. The hydrophobicity profile (top) of Coenzyme A Synthase was generated by TMpred program (Hofmann & Stoffel, 1993). A Modular representation of Coenzyme A Synthase is presented in the middle (Zhyvoloup, et al., 2003).|none]]<BR> | |||
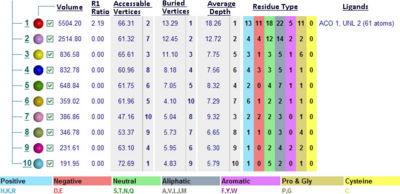
===Functional Sites Found By Pattern Search=== | |||
A motif database search (Bairoch, Bucher, & Hofmann, 1997) found a number of patterns in the ''Mus musculus'' Coenzyme A Synthase sequence ([[COASY_results#Functional Sites Found By Pattern Search|Table 1]]). Whilst some were to short to accurately determine if they were present in the sequence, the ATP/GTP-binding site motif A (P-loop) was found to be located on a known ligand binding location ([[COASY_results#Structure of Coenzyme A Synthase|Figure 2]]) which agreed with previous predicitions made for the DPCK domain ATP binding location ([[COASY_results#Structural Elements and Functional Binding Sites of Coenzyme A Synthase|Figure 5]], [[COASY_results#Electrostatic Surface Potential|Figure 6]]). <BR> | |||
====Table 1==== | |||
{| cellpadding="5" | |||
|+ '''PROSITE Motif search (Bairoch, Bucher, & Hofmann, 1997) results on Coenzyme A Synthase: ''Mus musculus''.''' | |||
|- | |- | ||
|- | |- | ||
| Line 98: | Line 159: | ||
|} | |} | ||
<BR> | |||
===Functional Sites Found by Sequence Conservation In Structurally Related Proteins=== | |||
A sequence identity conservation analysis of a structural alignment (Russell & Barton, 1992) of structurally related proteins (Holm & Sander, 1993) with ''Mus musculus'' CoAsy (PDB 2F6R) found that a number of residues outside the core of the structure were moderately preserved. Additionally, residues around the first helix, in particular the glycines and serines indicated to be binding sites in [[COASY_results#Structure of Coenzyme A Synthase|Figure 2]] (residues 70- 76), are highly conserved throughout the other structurally related proteins. The arginine on the eighth helix (residue 204), also indicated to be a ligand binding site ([[COASY_results#Structure of Coenzyme A Synthase|Figure 2]]), is highly conserved as well [[COASY_results#Functional Sites Found by Sequence Conservation In Structurally Related Proteins|(Figure 10)]]. | |||
[[Image:COASYSeqcongif.gif|framed|'''Figure 10 '''<BR> Functional Sites Found by Sequence Conservation In Structurally Related Proteins.|none]]<BR> | |||
<BR> | |||
===Functional Sites Found by Structure Conservation In Structurally Related Proteins=== | |||
An alignment of structurally conserved features for the found structurally related proteins on Coenzyme A Synthase (Holm & Sander, 1993) indicates that only the structural features located on the DPCK domain closest to the PPAT domain were structurally conserved. Helices three, four and five ([[COASY_results#Structure of Coenzyme A Synthase|Figure 2]]) were amongst the least structurally conserved whilst the area around the first helix including the predicted P-Loop location ([[COASY_results#Functional Sites Found By Pattern Search|Table 1]]) and the third strand are the strongest conserved along with other features located close to the core of CoAsy ([[COASY_results#Functional Sites Found by Structure Conservation In Structurally Related Proteins|Figure 11]] and [[COASY_results#Functional Sites Found by Structure Conservation In Structurally Related Proteins|Figure 12]]). | |||
[[Image:COASYStructcongif.gif|framed|'''Figure 11 '''<BR> Ribbon representation of ''Mus musculus'' Coenzyme A Synthase (RCSB, 2007) with conservation of structural features marked for structurally related proteins. Note P-Loop conservation (residues 65-85).|none]]<BR> | |||
[[Image:COASYStructureconservationstructurealignment.JPG|thumb|1050px|'''Figure 12 '''<BR> Structural feature conservation alignment of structurally related proteins for 2f6r:A CoAsy (Blue = more conserved).|none]]<BR> | |||
<BR> | |||
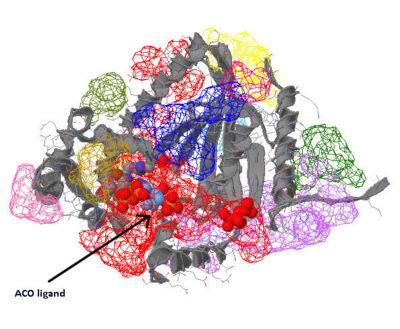
===Functional Sites Found Through ACO Crystallization=== | |||
The red binding cleft ([[COASY_results#Functional Sites Found Through ACO Crystallization|Figure 13]]) which the ACO ligand crystallised in 2F6R threads through the hole in the ACO cleft indicated in [[COASY_results#Electrostatic Surface Potential|Figure 6]], wrapping around CoAsy. This binding cleft also contains the predicted P-Loop region which ATP binds to ([[COASY_results#Structural Elements and Functional Binding Sites of Coenzyme A Synthase|Figure 5]]). EMBL EBI, 2005, predicts that ACO binds to residues 128, 219, 150 and 101 (Located on inner edges of helices surrounding location of ACO in [[COASY_results#Structural Elements and Functional Binding Sites of Coenzyme A Synthase|Figure 5]]). All of these sites are indicated as being ligand binding sites ([[COASY_results#Structure of Coenzyme A Synthase|Figure 2]]), although none of them are conserved, neither by structure ([[COASY_results#Functional Sites Found by Structure Conservation In Structurally Related Proteins|Figure 11]]) nor sequence ([[COASY_results#Functional Sites Found by Sequence Conservation In Structurally Related Proteins|Figure 10]]), across the other structurally related proteins. | |||
{| cellpadding="0" | border="1" | |||
|+ '''Figure 13''' | |||
|- | |||
|- | |||
|[[Image:COASYClefts.jpg|thumb|400px|none]] | |||
|- | |||
|[[Image:COASYCleft_table.jpg|thumb|400px|<BR> Location of binding clefts on Coenzyme A Synthase Mus. musculus with ACO and an unknown ligand (UNL).|none]] | |||
|- | |||
|} | |||
==Multiple Sequence Alignment== | |||
The multiple sequence alignment (msa), seen in [[COASY_results#Multiple Sequence Alignment|Figure 14]], shows slightly higher sequence similarity between the non-metazoan and the metazoan species in the DPCK region, but not in the PPAT region. The DPCK region of the human protein is shown as region 389-595 in the msa. The metazoan and non-metazoan species are slightly better aligned in the msa region 418-440, which is within the DPCK domain and the ATP binding site. This region of alignment appears to be the only well aligned segment between the metazoans and non-metazoans. However, the metazoan species have a high level of intra-species sequence similarity, which is particularly predominant within the mammalian species. | |||
Similarly, the ACO binding site (msa region 469-478) in the DPCK domain shows a high level of sequence conservation in the metazoans, but this region of sequence is not conserved in the bacterial species and is only slightly conserved in the other non-metazoans. In particular, the arginine at position 469 is fully conserved in the metazoans and ''Synechococcus sp'', ''Paramecium tetraurelia'' and ''Cryptococcus neoformans''. | |||
The multiple sequence alignment quality was excellent as the metazoan species showed high intra-sequence similarity. Although the non-metazoan species did not align well with the other sequences, this was due to sequence differences rather than poor alignment by the software program. | |||
[[Image:MSAfigure2.jpg|thumb|800px|'''Figure 14 '''<BR> Section of multiple sequence alignment showing the start of the P-loop (ATP binding site)and some conserved residues (left) and the ACO binding site (right)|none]]<BR> | |||
''' | |||
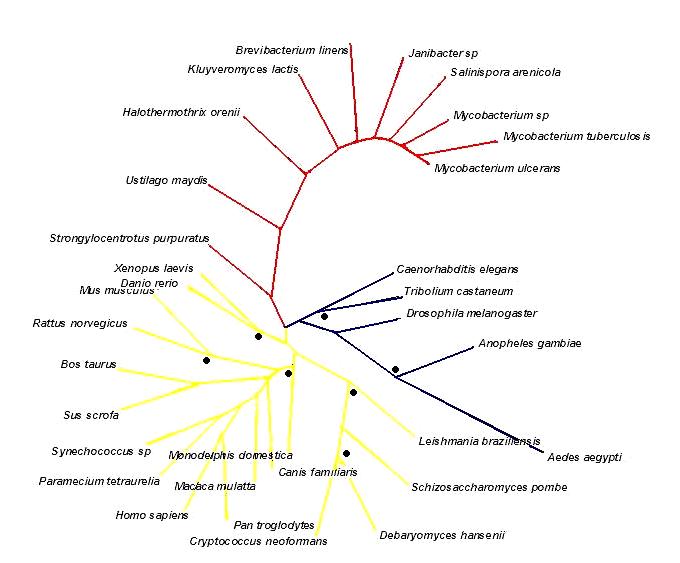
== | ===Tree=== | ||
There are three distinct branches present in the tree ([[COASY_results#Tree|Figure 15]]). The yellow branch consists of the mammals, the yeasts, some protozoan species, a cyanobacterium, a frog species and a species of zebra fish. The blue branch is composed of mosquitoes, a fruit fly species, a beetle species and worm species. The third red branch is predominantly bacterial with a sea urchin species included. | |||
This indicates conservation of CoAsy throughout the mammals and some of the higher eukaryotes. However, it is not clear as to why a protozoan or a cyanobacterium is present in the yellow branch, but this must indicate sequence conservation of human CoAsy in these species. Similarly, it is contradictory that there is a separate branch for the mosquitoes, flies, worms and beetles as these seem more likely on an evolutionary basis to have a more similar CoAsy sequence to mammals than any protozoan or bacterial species. However, the tree does indicate that there must be some sequence divergence from human CoAsy in the blue branch species. From examination of the branch positions, it can be ascertained that the species in the yellow and blue branches are more closely related to each other than to the red branch. It is evident that bacterial species on the whole have low sequence similarity to metazoan species as the red branch shows a strong level of divergence from both the blue and the yellow branches. | |||
The tree has a varied selection of organisms from many phyla, which gives good ground for comparison between species. However, the addition of more protozoan and yeast species would confirm their placement on the tree. Similarly, more fly, worm and beetle species would be beneficial to ascertaining their position as a separate branch on the tree. | |||
[[Image:phylogenic_tree3.jpg|thumb|800px|'''Figure 15 '''<BR> Phylogenetic tree. The dots are the bootstrapped values over 75%|none]]<BR> | |||
Latest revision as of 23:25, 10 June 2007
Structure of Coenzyme A Synthase
Conezyme A Synthase is structurally composed of seven strands, eleven helices and thirteen beta turns (EMBL EBI, 2005) (see Figure 2). Analysis of structurally related proteins (Holm & Sander, 1993) showed a trend for transferase (RCSB, 2007) class proteins with Rossmann class folds (Rossmann, 1973). The Rossmann topologies of folds are an alpha-beta class fold that forms a three or more layer beta strand sandwich alternating with alpha helices (beta-alpha-beta-alpha-beta). PFAM classification placed these into either the Cytidylyltransferase or Dephospho-CoA kinase familes, matching the two domains of Coenzyme A Synthase. Those classified under the Dephospho-CoA kinase type were commonly of the P-loop containing nucleotide triphosphate hydrolases (Sanger Institute, 2005) homology whilst those classified as Cytidylyltransferase were of the Tyrosol-Transfer RNA Synthetase (Sanger Institute, 2005). As the majority of the structurally related proteins identified contained the DPCK domain solely, and the motif for a P-loop (Table 1) was identified in the Conzyme A Synthase sequence (Bairoch, Bucher, & Hofmann, 1997) it is suggested that Coenzyme A Synthase is also of the P-loop containing nucleotide triphosphate hydrolases homology of folds.
Localisation Expression of Coenzyme A Synthase
Baseline expression of CoAsy was present in all tissues. Expression was slightly elevated in adipose tissue, and highly elevated in the liver and kidney (See Figure 3). Baseline expression of the other enzymes involved in the CoAsy pathway was also present in all tissues. However, expression of these enzymes was otherwise unlike that of CoA, as they were not particularly elevated in the liver and kidney (data not shown).
Domain and Structural Analysis
Local sequence alignment showed that CoAsy Chain A residues 13-281 corresponded to residues 295-563 on full length CoAsy. This indicates that a 295 amino acid stretch extending from residues 1 to 295 on the full length protein was cut out during structural analysis and was not present on chain A.
Bioinformatic analysis using NCBI Entrez protein indicated that mouse CoAsy (full length) possess two domains, a PPAT and a DPCK domain. The PPAT domain extended from residues 194-338, and the DPCK domain from residues 359-536. PFAM analysis of CoAsy chain A also indicated the presence of two domains, the DPCK domain from residues 64-242 and a CTP transferase 2 domain (part of the cytidylyltransferase family, of which PPAT is a member) extending from residues 1-44, which corresponds to a fragment of the PPAT domain. The position of these domains on CoAsy chain A is shown in Figure 4. Profunc analysis of CoAsy chain A detected the DPCK domain, but not the fragment of the PPAT domain.
Structural Elements and Functional Binding Sites of Coenzyme A Synthase
Comparison to CoAsy’s closest structurally related protein, Dephospho-CoenzymeA Kinase, finds that whilst it shares only twenty percent of its sequence, it is structurally very similar (Figure 5). Apart from the lack of a PPAT domain on the Dephospho-CoenzymeA Kinase molecule (see Figure 4 for comparative location of PPAT), all structural features are roughly the same between the two proteins. As helices, sheets and loops stay relatively constant between the two, and Dephospho-CoenzymeA Kinase has a related function to CoAsy it can be assumed that the location of the ATP binding site on the DPCK domain of CoAsy is the same as that on Dephospho-CoenzymeA Kinase.
Electrostatic Surface Potential
Analysis of electrostatic surface potential maps for CoAsy finds that there is a strongly positive charged region, between residues 70 and 80, on the first helix that lines up with the proposed ATP binding site (Figure 6). It can also be noted that the inner wall of the major cleft alternates strongly between a positive and negative charge and this lines up with the ACO ligand that is included in the 2F6R PDB entry at this location (EMBL EBI, 2005).
Residue Flexibility
Structural analysis of Coenzyme A Synthase B-Factor scores finds it to be most stable close to the center, and on the N-terminal of the protein. The most flexible regions are on the eighth helix and on the outer edges of the protein (Figure 7). The position of most flexibility on the protein is at the top of the eight loop, on the DPCK domain, and this lies between both the bind site for ATP and the location of ACO on the 2F6R PDB entry (EMBL EBI, 2005).
Hydrophobicity
Hydrophobic mapping of Mus musculus Coenzmye A Synthase finds a number of hydrophobic stretches including the inner strands and the N terminal Figure 8. A study by (Zhyvoloup, et al., 2003) also found that the Mus musculus CoAsy had a hydrophobic N terminal and that this is a conserved feature for CoAsy across a broad number of species Figure 9.
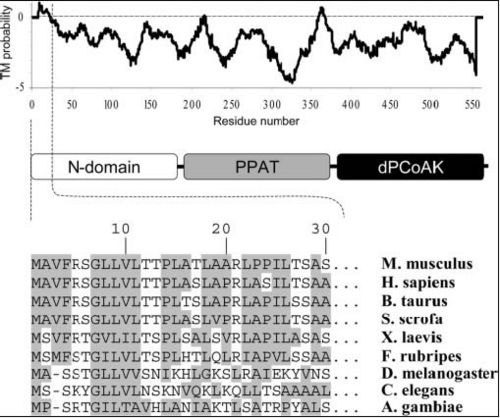
Sequence alignment of N domain (first 30 residues) of Coenzyme A Synthase proteins (bottom) with hydrophobic residues shaded. The hydrophobicity profile (top) of Coenzyme A Synthase was generated by TMpred program (Hofmann & Stoffel, 1993). A Modular representation of Coenzyme A Synthase is presented in the middle (Zhyvoloup, et al., 2003).
Functional Sites Found By Pattern Search
A motif database search (Bairoch, Bucher, & Hofmann, 1997) found a number of patterns in the Mus musculus Coenzyme A Synthase sequence (Table 1). Whilst some were to short to accurately determine if they were present in the sequence, the ATP/GTP-binding site motif A (P-loop) was found to be located on a known ligand binding location (Figure 2) which agreed with previous predicitions made for the DPCK domain ATP binding location (Figure 5, Figure 6).
Table 1
| Pattern-ID: | GLYCOSAMINOGLYCAN PS00002 PDOC00002 |
| Pattern-DE: | Glycosaminoglycan attachment site |
| Pattern: | SG.G |
| Position: | 84 SGSG |
| Pattern-ID: | PKC_PHOSPHO_SITE PS00005 PDOC00005 |
| Pattern-DE: | Protein kinase C phosphorylation site |
| Pattern: | [ST].[RK] |
| Position: | 3 SDK |
| Position: | 52 SFR |
| Position: | 86 SGK |
| Pattern-ID: | CK2_PHOSPHO_SITE PS00006 PDOC00006 |
| Pattern-DE: | Casein kinase II phosphorylation site |
| Pattern: | [ST].{2}[DE] |
| Position: | 39 SHNE |
| Position: | 251 TLWE |
| Position: | 260 SQVE |
| Pattern-ID: | MYRISTYL PS00008 PDOC00008 |
| Pattern-DE: | N-myristoylation site |
| Pattern: | G[^EDRKHPFYW].{2}[STAGCN][^P] |
| Position: | 82 GISGSG |
| Position: | 222 GLSEAA |
| Pattern-ID: | ATP_GTP_A PS00017 PDOC00017 |
| Pattern-DE: | ATP/GTP-binding site motif A (P-loop) |
| Pattern: | [AG].{4}GK[ST] |
| Position: | 82 GISGSGKS |
| Pattern-ID: | UPF0038 PS01294 PDOC00996 |
| Pattern-DE: | Uncharacterized protein family UPF0038 signature |
| Pattern: | G.[LI].R.{2}L.{4}F.{8}[LIV].{5}P.[LIV] |
| Position: | 136 GTINRKVLGSRVFGNKKQMKILTDIVWPVI |
Functional Sites Found by Sequence Conservation In Structurally Related Proteins
A sequence identity conservation analysis of a structural alignment (Russell & Barton, 1992) of structurally related proteins (Holm & Sander, 1993) with Mus musculus CoAsy (PDB 2F6R) found that a number of residues outside the core of the structure were moderately preserved. Additionally, residues around the first helix, in particular the glycines and serines indicated to be binding sites in Figure 2 (residues 70- 76), are highly conserved throughout the other structurally related proteins. The arginine on the eighth helix (residue 204), also indicated to be a ligand binding site (Figure 2), is highly conserved as well (Figure 10).
Functional Sites Found by Structure Conservation In Structurally Related Proteins
An alignment of structurally conserved features for the found structurally related proteins on Coenzyme A Synthase (Holm & Sander, 1993) indicates that only the structural features located on the DPCK domain closest to the PPAT domain were structurally conserved. Helices three, four and five (Figure 2) were amongst the least structurally conserved whilst the area around the first helix including the predicted P-Loop location (Table 1) and the third strand are the strongest conserved along with other features located close to the core of CoAsy (Figure 11 and Figure 12).
Functional Sites Found Through ACO Crystallization
The red binding cleft (Figure 13) which the ACO ligand crystallised in 2F6R threads through the hole in the ACO cleft indicated in Figure 6, wrapping around CoAsy. This binding cleft also contains the predicted P-Loop region which ATP binds to (Figure 5). EMBL EBI, 2005, predicts that ACO binds to residues 128, 219, 150 and 101 (Located on inner edges of helices surrounding location of ACO in Figure 5). All of these sites are indicated as being ligand binding sites (Figure 2), although none of them are conserved, neither by structure (Figure 11) nor sequence (Figure 10), across the other structurally related proteins.
Multiple Sequence Alignment
The multiple sequence alignment (msa), seen in Figure 14, shows slightly higher sequence similarity between the non-metazoan and the metazoan species in the DPCK region, but not in the PPAT region. The DPCK region of the human protein is shown as region 389-595 in the msa. The metazoan and non-metazoan species are slightly better aligned in the msa region 418-440, which is within the DPCK domain and the ATP binding site. This region of alignment appears to be the only well aligned segment between the metazoans and non-metazoans. However, the metazoan species have a high level of intra-species sequence similarity, which is particularly predominant within the mammalian species. Similarly, the ACO binding site (msa region 469-478) in the DPCK domain shows a high level of sequence conservation in the metazoans, but this region of sequence is not conserved in the bacterial species and is only slightly conserved in the other non-metazoans. In particular, the arginine at position 469 is fully conserved in the metazoans and Synechococcus sp, Paramecium tetraurelia and Cryptococcus neoformans.
The multiple sequence alignment quality was excellent as the metazoan species showed high intra-sequence similarity. Although the non-metazoan species did not align well with the other sequences, this was due to sequence differences rather than poor alignment by the software program.
Tree
There are three distinct branches present in the tree (Figure 15). The yellow branch consists of the mammals, the yeasts, some protozoan species, a cyanobacterium, a frog species and a species of zebra fish. The blue branch is composed of mosquitoes, a fruit fly species, a beetle species and worm species. The third red branch is predominantly bacterial with a sea urchin species included.
This indicates conservation of CoAsy throughout the mammals and some of the higher eukaryotes. However, it is not clear as to why a protozoan or a cyanobacterium is present in the yellow branch, but this must indicate sequence conservation of human CoAsy in these species. Similarly, it is contradictory that there is a separate branch for the mosquitoes, flies, worms and beetles as these seem more likely on an evolutionary basis to have a more similar CoAsy sequence to mammals than any protozoan or bacterial species. However, the tree does indicate that there must be some sequence divergence from human CoAsy in the blue branch species. From examination of the branch positions, it can be ascertained that the species in the yellow and blue branches are more closely related to each other than to the red branch. It is evident that bacterial species on the whole have low sequence similarity to metazoan species as the red branch shows a strong level of divergence from both the blue and the yellow branches.
The tree has a varied selection of organisms from many phyla, which gives good ground for comparison between species. However, the addition of more protozoan and yeast species would confirm their placement on the tree. Similarly, more fly, worm and beetle species would be beneficial to ascertaining their position as a separate branch on the tree.
Abstract | Introduction | Results | Discussion | Conclusion | Method | References