Arylformamidase Structure: Difference between revisions
No edit summary |
|||
| (27 intermediate revisions by the same user not shown) | |||
| Line 13: | Line 13: | ||
== '''Results''' == | |||
== Structure of Arylformamidase == | == Structure of Arylformamidase == | ||
Structure was determined using X-ray diffraction by the Joint Center for Structural Genomics (JCSG). The organism is Silicibacter SP. TM1040 and the protein expression system is Escherichia Coli (vector type: plasmid). The resolution is 1.79 A with R-value of 0.224 and R-free value of 0.270. The closer the R values are to each other, the better the quality of the structure. | |||
| Line 25: | Line 29: | ||
''The image above shows the chains A (upper right), B (upper left), C (lower right) & D (lower left) interacting. The molecules in the middle of chains A & B and chains C & D is phosphate ion (PO4). The green molecule between chain B & D is a magnesium ion (Mg). These ions aren't biologically significant and could only be an artefact. When crystallizing proteins they often form complexes (dimer, tetramers etc) but that doesn't mean that the functional structure is the same. They could be functional monomers. | ''The image above shows the chains A (upper right), B (upper left), C (lower right) & D (lower left) interacting. The molecules in the middle of chains A & B and chains C & D is phosphate ion (PO4). The green molecule between chain B & D is a magnesium ion (Mg). These ions aren't biologically significant and could only be an artefact. When crystallizing proteins they often form complexes (dimer, tetramers etc) but that doesn't mean that the functional structure is the same. They could be functional monomers. The chains in the protein of interest exist as individual functional units because in the PDB file it assumes the functional biological molecule as a monomer.'' | ||
''Image from PDB ProteinWorkshop 1.5'' | ''Image from PDB ProteinWorkshop 1.5'' | ||
| Line 42: | Line 47: | ||
'''''The protein backbone is coloured by conformation type:''''' | '''''The protein backbone is coloured by conformation type:''''' | ||
'''Turn - blue''' | '''''Turn - blue''''' | ||
'''Coil- pink''' | '''''Coil- pink''''' | ||
'''Helix- green''' | '''''Helix- green''''' | ||
'''strand- purple''' | '''''strand- purple''''' | ||
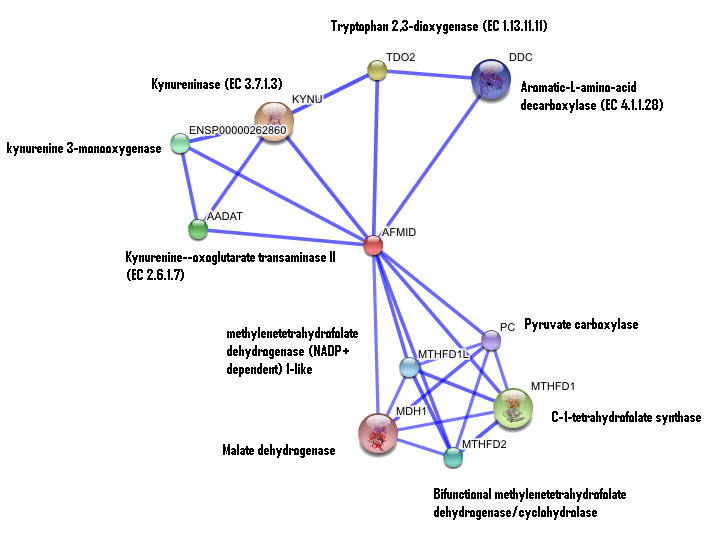
== '''Interaction of human arylformamidase (AFMID) with other proteins''' == | == '''Interaction of human arylformamidase (AFMID) with other proteins''' == | ||
| Line 56: | Line 62: | ||
The interaction between the proteins have been determined from curated STRING database (significant score). However there is no significant evidence for: | ''The interaction between the proteins have been determined from curated STRING database (significant score). However there is no significant evidence for:'' | ||
1- Neighborhood in the genome | ''1- Neighborhood in the genome'' | ||
2- Gene fusions | ''2- Gene fusions '' | ||
3- Cooccurence across genomes | ''3- Cooccurence across genomes '' | ||
4- Co-Expression | ''4- Co-Expression '' | ||
5- Experimental/Biochemical data | ''5- Experimental/Biochemical data'' | ||
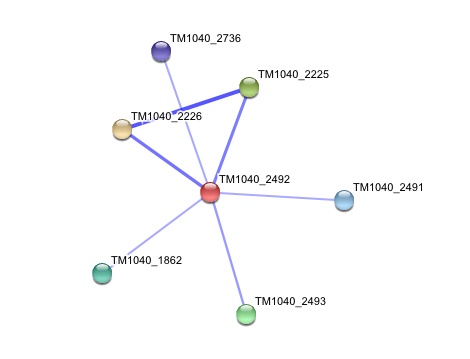
== '''Interaction of Silicibacter Sp. arylformamidase (AFMID) with other proteins''' == | |||
[[Image:Examplec.jpg]] | [[Image:Examplec.jpg]] | ||
| Line 87: | Line 94: | ||
There is no significant evidence for these interactions (score= ~0.5) | ''There is no significant evidence for these interactions (score= ~0.5)'' | ||
== '''DALI OUTPUT''' == | |||
| Line 101: | Line 108: | ||
'''Metagenomic Archea Carboxylesterase (Chain A ONLY)''' | '''Figure: Metagenomic Archea Carboxylesterase (Chain A ONLY)''' | ||
[[Image:ChainA 2c7b.PNG]] | [[Image:ChainA 2c7b.PNG]] | ||
[[Image:Carboxylase.txt ]] | [[Image:Carboxylase.txt ]] | ||
| Line 109: | Line 116: | ||
'''PDB''' [http://www.rcsb.org/pdb/explore/explore.do?structureId=2C7B link title] | '''PDB''' [http://www.rcsb.org/pdb/explore/explore.do?structureId=2C7B link title] | ||
Note: Chain B not shown | ''Note: Chain B not shown'' | ||
''From PDB ProteinWorkshop 1.5'' | |||
'''Archaeoglobus fulgidus Carboxylesterase (Chain A ONLY)''' | '''Figure: Archaeoglobus fulgidus Carboxylesterase (Chain A ONLY)''' | ||
[[Image:ChainA 1jji.PNG]] | [[Image:ChainA 1jji.PNG]] | ||
[[Image:Carboxylesterase (archaeon).txt]] | [[Image:Carboxylesterase (archaeon).txt]] | ||
| Line 120: | Line 129: | ||
'''PDB''' [http://www.rcsb.org/pdb/explore/explore.do?structureId=1JJI link title] | '''PDB''' [http://www.rcsb.org/pdb/explore/explore.do?structureId=1JJI link title] | ||
Note: Chains B, C & D not shown | ''Note: Chains B, C & D not shown'' | ||
''From PDB ProteinWorkshop 1.5'' | |||
Both of the above Archaeal carboxylesterases' chains exist as monomers (from literature). Hence it is expected that our protein exists as a monomer but during crystalization it interacts with its chains. | ''Both of the above Archaeal carboxylesterases' chains exist as monomers (from literature). Hence it is expected that our protein exists as a monomer but during crystalization it interacts with its chains.'' | ||
== '''Secondary structure analysis''' == | == '''Secondary structure analysis''' == | ||
| Line 135: | Line 147: | ||
'''Archeon Carboxylesterase secondary structure''' | '''Figure: Archeon Carboxylesterase secondary structure''' | ||
[[Image:Pdbsums archeal.PNG]] | |||
''The secondary structure shows the conservation of the order of different conformation types between the protein of interest and the archaeal carboxylesterases.'' | |||
''Images from PDBsum'' | |||
== '''The conservation of the ser/his/asp catalytic triad''' == | == '''The conservation of the ser/his/asp catalytic triad''' == | ||
| Line 178: | Line 194: | ||
[[Image:CATALYTIC TRIAD 1.PNG]] | [[Image:CATALYTIC TRIAD 1.PNG]] | ||
''The above image shows the distance between the catalytic triad conserved residues and how each amino acid is linked to a turn region.'' | ''The above image shows the distance between the catalytic triad conserved residues and how each amino acid is linked to a turn region. This catalytic triad is also conserved in the Metagenomic Archea Carboxylesterase (PDB ID 2C7B) and the Archaeoglobus fulgidus Carboxylesterase (PDB ID 1JJI)'' | ||
'' From PDB ProteinWorkshop 1.5'' | '' From PDB ProteinWorkshop 1.5'' | ||
'''Figure: The conserved catalytic triad in Archaeoglobus fulgidus Carboxylesterase (PDB ID 1JJI)''' | |||
[[Image:Cat triad 1jji.PNG]] | |||
''The catalytic triad in Archaeoglobus fulgidus Carboxylesterase is very close to the ligand which is also present in aryformamidase.'' | |||
'''Sequence alignment:''' | |||
''Sections of alignment showing the conserved residues across bacterial and eukaryotic species.'' | |||
[[Image:Alignment1.jpg]] | |||
[[Arylformamidase | Return to the main page...]] | [[Arylformamidase | Return to the main page...]] | ||
Latest revision as of 01:35, 10 June 2008
Methods
The structure of arylfromamidase was obtained from RCSB Protein Data Bank (PDB ID: 2PBL). http://www.rcsb.org/pdb/home/home.do
The predicted interaction arylformamidase with other proteins was determined using the STRING database (STRING: Search Tool for the Retrieval of Interacting Genes/Proteins). http://string.embl.de//
The DALI database was used for the structural comparison of arylformamidase with other proteins. http://ekhidna.biocenter.helsinki.fi/dali_server/
PDBsum database was used to determine the secondary structure of arylformamidase. http://www.ebi.ac.uk/pdbsum/
Results
Structure of Arylformamidase
Structure was determined using X-ray diffraction by the Joint Center for Structural Genomics (JCSG). The organism is Silicibacter SP. TM1040 and the protein expression system is Escherichia Coli (vector type: plasmid). The resolution is 1.79 A with R-value of 0.224 and R-free value of 0.270. The closer the R values are to each other, the better the quality of the structure.
Figure: Arylformamidase (All Chains)
The image above shows the chains A (upper right), B (upper left), C (lower right) & D (lower left) interacting. The molecules in the middle of chains A & B and chains C & D is phosphate ion (PO4). The green molecule between chain B & D is a magnesium ion (Mg). These ions aren't biologically significant and could only be an artefact. When crystallizing proteins they often form complexes (dimer, tetramers etc) but that doesn't mean that the functional structure is the same. They could be functional monomers. The chains in the protein of interest exist as individual functional units because in the PDB file it assumes the functional biological molecule as a monomer.
Image from PDB ProteinWorkshop 1.5
Figure:Chain A of arylformamidase
The red molecule in the middle is an unknown ligand containing a ring composed of 9 oxygen molecules. The green sphere is a chloride ion.
Image from PDB ProteinWorkshop 1.5
The protein backbone is coloured by conformation type:
Turn - blue
Coil- pink
Helix- green
strand- purple
Interaction of human arylformamidase (AFMID) with other proteins
The interaction between the proteins have been determined from curated STRING database (significant score). However there is no significant evidence for:
1- Neighborhood in the genome
2- Gene fusions
3- Cooccurence across genomes
4- Co-Expression
5- Experimental/Biochemical data
Interaction of Silicibacter Sp. arylformamidase (AFMID) with other proteins
TM1040_2226 Tryptophan 2,3-dioxygenase (279 aa)
TM1040_2225 Kynureninase (396 aa)
TM1040_2493 Succinic semialdehyde dehydrogenase (490 aa)
TM1040_1862 Hypothetical protein (212 aa)
TM1040_2491 Creatinase (402 aa)
TM1040_2736 Transketolase, putative (794 aa)
There is no significant evidence for these interactions (score= ~0.5)
DALI OUTPUT
The DALI tool produces proteins that are structurally similar to the protein of interest.
The search result showed similarities to mostly carboxylesterases/hydrolases. Hence there is strong evidence that our protein might also be a carboxylesterase.
Figure: Metagenomic Archea Carboxylesterase (Chain A ONLY)
PDB link title
Note: Chain B not shown
From PDB ProteinWorkshop 1.5
Figure: Archaeoglobus fulgidus Carboxylesterase (Chain A ONLY)
File:Carboxylesterase (archaeon).txt
PDB link title
Note: Chains B, C & D not shown
From PDB ProteinWorkshop 1.5
Both of the above Archaeal carboxylesterases' chains exist as monomers (from literature). Hence it is expected that our protein exists as a monomer but during crystalization it interacts with its chains.
Secondary structure analysis
PDBSum output for arylformamidase
PDBSUM [1]
Figure: Archeon Carboxylesterase secondary structure
The secondary structure shows the conservation of the order of different conformation types between the protein of interest and the archaeal carboxylesterases.
Images from PDBsum
The conservation of the ser/his/asp catalytic triad
Yellow indicates conservation
Blue indicates semi-conservation
Figure: The catalytic triad
The above image shows the conserved residues of the catalytic triad in arylformamidase, with the unknown ligand (Blue) protruding from a surface groove. The residues are serine 136, Histidine 241 and Glutamate 214. Note: The actual residue numbers are n+1
Image generated using Pymol
Figure: The conserved residues of arylformamidase
The blue region shows the residues conserved among species. It is mostly around the unknown ligand. The conserved residues were obtained from observing the clustal alignment.
Image generated using Pymol
Figure: The catalytic triad
The above image shows the distance between the catalytic triad conserved residues and how each amino acid is linked to a turn region. This catalytic triad is also conserved in the Metagenomic Archea Carboxylesterase (PDB ID 2C7B) and the Archaeoglobus fulgidus Carboxylesterase (PDB ID 1JJI)
From PDB ProteinWorkshop 1.5
Figure: The conserved catalytic triad in Archaeoglobus fulgidus Carboxylesterase (PDB ID 1JJI)
The catalytic triad in Archaeoglobus fulgidus Carboxylesterase is very close to the ligand which is also present in aryformamidase.
Sequence alignment:
Sections of alignment showing the conserved residues across bacterial and eukaryotic species.