Hypothetical protein Discussion: Difference between revisions
No edit summary |
No edit summary |
||
| Line 6: | Line 6: | ||
Structural alignments found that the query protein matched well against RbsD transporters, however there were no FucU proteins to compare it to. This caused a limit to the structural comparisons that were made. From the structural alignments it can be assumed that the query forms a decamerichal oligomer possibly containing putative chloride binding motif involved with polymerisation. These theories are based on the assumption that there is no significant difference between tertiary structures between FucU and RbsD. This is a valid assumption because the secondary structures are similar. | Structural alignments found that the query protein matched well against RbsD transporters, however there were no FucU proteins to compare it to. This caused a limit to the structural comparisons that were made. From the structural alignments it can be assumed that the query forms a decamerichal oligomer possibly containing putative chloride binding motif involved with polymerisation. These theories are based on the assumption that there is no significant difference between tertiary structures between FucU and RbsD. This is a valid assumption because the secondary structures are similar. | ||
The 1ogd binding pocket (RsbD protein found in ''B.Subtillius'') consists of, D-28, Y-120, N-122, H-98 and K-102. It also contained a H-20 from a neighboring monomer. The 2ob5A binding pocket is similar it has a corosponding H-22 from the other protein as well as matching D-30 and Y-136 | The 1ogd binding pocket (RsbD protein found in ''B.Subtillius'') consists of, D-28, Y-120, N-122, H-98 and K-102. It also contained a H-20 from a neighboring monomer. The 2ob5A binding pocket is similar it has a corosponding H-22 from the other protein as well as matching D-30 and Y-136. There was no corresponding N-122, possibly due to a deletion. The H-98 and The K-102 on the other hand were substituted with R-114 and Y-118 respectively. | ||
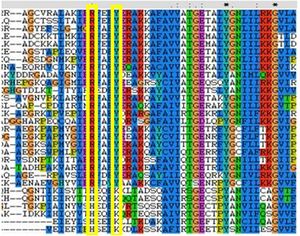
The R and Y substitutions has been shown to be highly conserved in all FucU proteins as seen in the MSA (see figure). | The R and Y substitutions has been shown to be highly conserved in all FucU proteins as seen in the MSA (see figure). | ||
[[Image:conres.jpg|right|thumbnail|Figure 1. This MSA shows the binding redidues that have been substituted in RbsD and FucU]] | [[Image:conres.jpg|right|thumbnail|Figure 1. This MSA shows the binding redidues that have been substituted in RbsD and FucU]] | ||
Revision as of 00:12, 16 June 2009
The hypothetical putative sugar binding protein 2ob5 is part of the RbsD/FucU superfamily in particular it appears to be a FucU protein that catalysis the anomeric change of alpha-purine-fucose into beta-purine-fucose. This was found through combination of evolutionary, structural and functional analysis, this was all compared to existing literature.
The MSA and tree diagrams found a significantly divergent group of sequences that was completely separate from the remainder of the other groups (green, bootstrap 100). This divergent group was attributed to being RbsD protein transporters. It is hypothesized that the gene encoding the RbsD transporter had undergone duplication in a common ancestor, as indicated by the presence of both RbsD and FucU(which are highly divergent) in the bacterium E. coli. It is believed that this split ultimately created the FucU paralog. Our query protein was found to have higher similarity to the FucU group of proteins. Furthermore, we did not encounter any RbsD transporter proteins in eukaryotes.
The query protein was found in several alpha-proteobacteria (high similarity), supported by a bootstrap value of 100 (tree, blue). The chordate group was monophyletic (red, bootstrap value of 99), but sister group to several bacteria (yellow, bootstrap value of 75). We could not find any significant differences distinguishing these bacteria from the bacteria that were not sister group to chordates (purple).This raises a question about the evolution of our query protein: why do some bacteria have transporters that are more similar to eukaryotic proteins than to other bacteria?
Structural alignments found that the query protein matched well against RbsD transporters, however there were no FucU proteins to compare it to. This caused a limit to the structural comparisons that were made. From the structural alignments it can be assumed that the query forms a decamerichal oligomer possibly containing putative chloride binding motif involved with polymerisation. These theories are based on the assumption that there is no significant difference between tertiary structures between FucU and RbsD. This is a valid assumption because the secondary structures are similar. The 1ogd binding pocket (RsbD protein found in B.Subtillius) consists of, D-28, Y-120, N-122, H-98 and K-102. It also contained a H-20 from a neighboring monomer. The 2ob5A binding pocket is similar it has a corosponding H-22 from the other protein as well as matching D-30 and Y-136. There was no corresponding N-122, possibly due to a deletion. The H-98 and The K-102 on the other hand were substituted with R-114 and Y-118 respectively. The R and Y substitutions has been shown to be highly conserved in all FucU proteins as seen in the MSA (see figure).
This further suports the fact that the query protein is a member of the FucU group of transporters. Previous studies have reiterated this point, this change in the binding pocket possibly accounts for the difference in functions between the paralogs.
The crystallographic structure of 1ogD contains a metal-ion coordinating pocket formed by the interface between subunits adjacent between upper and lower sections of the ring. This ion – potentially a Cl- - is cocrystallised in the structure of 1ogD, and appears to be coordinated by the Lys2 residue. This metal ion would have a potential stabilising effect on the oligomer. Visual characteristics of the corresponding 2ob5 region suggest that 2ob5 also exists as a decamer. However, the chloride-coordinating Lysine is replaced by an aliphatic leucine in 2ob5 and all other fucose-binding proteins analysed (complete alignment). From this, we hypothesise that the chloride ion is absent in fucose-binding proteins, but that the proteins are still able to assume a stable decameric ring oligomer.
The functional analysis's completed failed to give any major insite in to the function of the target protein. The sting analsyis only showed three possible interacting proteins, all of the interactions were not very conclusive and failed to tell us anything about the protein. The Profunc results reconfirmed the fact that the target protein belonged to the FucU and RbsD superfamily. Unfortunally there was only two other members of this group that have their structures crystallized and both are members of the RbsD group of transporters. This lack of homologes caused the profunc results to be inconclusive in all aspects.
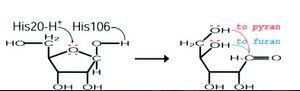
All information regarding the function of these proteins was gained from previous studies. The RbsD acts as a mutarotase to convert beta purine to beta furine. A proposed mode of action for this role of RbsD mutarotase can be seen in figure 2.
The His20 residue protonates the O5 and the His106 deprotonates the OH attached to the C1. This causes ring-breakage allowing the formation of an equilibrium between the furan and pyran forms. The FucU is fundamentally different, changing the beta to alpha pyranse. The altered FucU residues in the binding site are thought to be the reason for the different functions. The two residues may cause the conversion of L-fucose by associating with the OH group at C-1 position of the sugar.
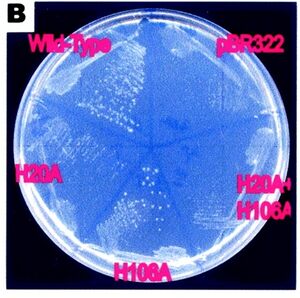
The importance of the H-20 and H-106 residues in the function of RsbD has been shown in an expereiment where these residues have been substituted for alanene. The Bacteria without H-20 residue had no pyrenase activity, and the residues without the H-106 lost appoximitly half of their activity. This was shown by growing them in a ribose substrate where the pressence of RsbD is necessary for the cell to survive (see figure 3)
Previous research has shown that although RbsD did not show any activity for L-fucose as a substrate FucU exhibited a limited pyranase activity for D-ribose. This is expected as the FucU protein still contains the functionaly important H-20 redsidue.
Abstract | Introduction | Method |
Results | Discussion | Conclusion | References