Discussion of SNAPG: Difference between revisions
No edit summary |
No edit summary |
||
| Line 5: | Line 5: | ||
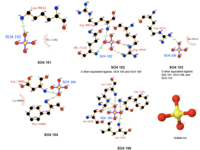
[[Image:Sulfate ion ligand interactions.png|thumb|200px|right|Figure 2. Various interaction of sulfanate ions]] | [[Image:Sulfate ion ligand interactions.png|thumb|200px|right|Figure 2. Various interaction of sulfanate ions]] | ||
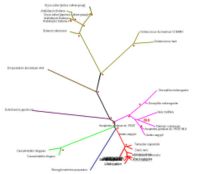
[[Image:Rad.JPG|thumb|200px|Figure 3. Evolutionary analysis of the tree with radial view. Red dotted signs means bootstrap value more than 75% or 75.00|right]] | |||
nanti aku mau jelasin ttg ligandnya | nanti aku mau jelasin ttg ligandnya | ||
Revision as of 11:47, 11 June 2007

nanti aku mau jelasin ttg ligandnya
Tea: evolution linked with function.
SNAP-gamma protein evolutionary tree mostly contains more than 75% boostrapvalue. Thus, red dot signs was assign to represent more than 75% boostrap value. The value was indicated the degree of confidence about the branching order. As an example: in the diagram phylogenetic tree (see Figure 3), there is 39.0 boostrap value in branching Apis melifera and Tribolium castaneum. The boostrap value is below 75%, thus it could be concluded that the degree of confidence of branching tree in the correct order is far below 95%. On the other hand, boostrap value more than 75% or 75.00 means 95% confidence that the branching tree is in the correct order
As indicated in diagram above, SNAP-gamma protein was evolved in eukaryotes. There is no prokaryotes detected in the phylogenetic tree.
Function (in various organism) = nunggu justine
Thus, it could be concluded that SNAP-gamma protein is important protein in eukaryotes animal.