ATP binding domain 4 Structures: Difference between revisions
From MDWiki
Jump to navigationJump to search
Anharmustafa (talk | contribs) No edit summary |
Anharmustafa (talk | contribs) No edit summary |
||
| Line 2: | Line 2: | ||
== Useful info == | == '''Useful info''' == | ||
Here is some useful paper. check it out | Here is some useful paper. check it out | ||
| Line 9: | Line 9: | ||
Interpro: http://www.ebi.ac.uk/interpro/DisplayIproEntry?ac=IPR002761 | Interpro: http://www.ebi.ac.uk/interpro/DisplayIproEntry?ac=IPR002761 | ||
== SCOP Work == | == '''SCOP Work''' == | ||
http://scop.mrc-lmb.cam.ac.uk/scop/data/scop.b.d.cj.c.b.bb.html | http://scop.mrc-lmb.cam.ac.uk/scop/data/scop.b.d.cj.c.b.bb.html | ||
| Line 15: | Line 15: | ||
== Pymol Work == | == '''Pymol Work''' == | ||
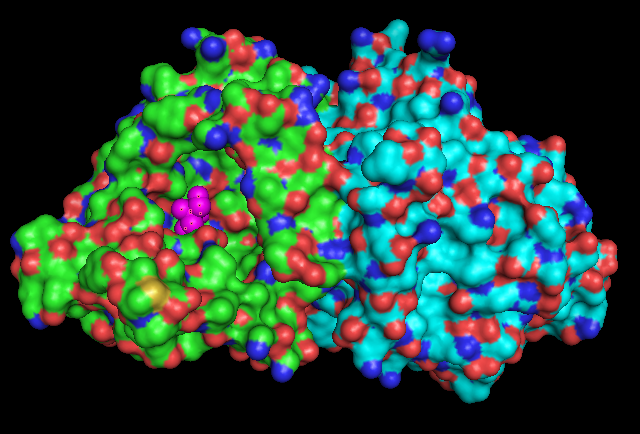
Surface Structure | |||
== Surface Structure == | |||
[[Image:1ru8.png]] | [[Image:1ru8.png]] | ||
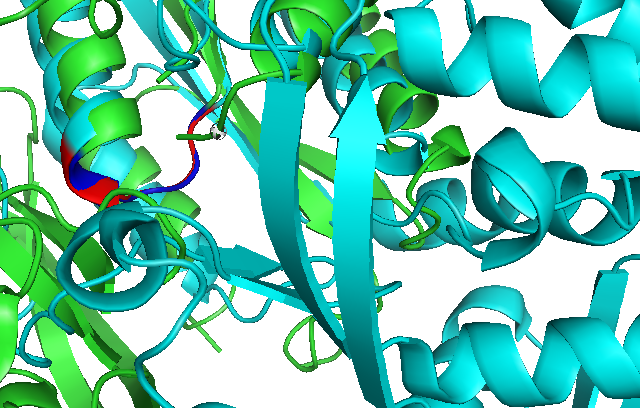
Secondary Structure and Location of P-loop (ATP binding site) | |||
== Secondary Structure and Location of P-loop (ATP binding site) == | |||
[[Image:PDBsum chain.JPG]] | [[Image:PDBsum chain.JPG]] | ||
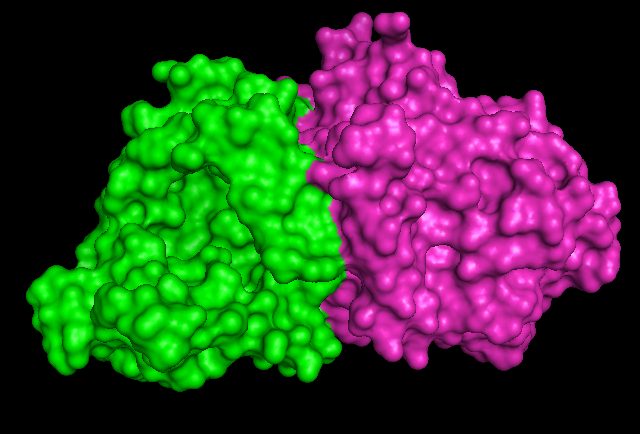
1RU8 has 2 domain | |||
== 1RU8 has 2 domain == | |||
[[Image:1RU8 as 2 domain.png]] | [[Image:1RU8 as 2 domain.png]] | ||
Electrostatic Surface Potential | |||
== Electrostatic Surface Potential == | |||
[[Image:1ru8 electro surface.png]] | [[Image:1ru8 electro surface.png]] | ||
| Line 35: | Line 39: | ||
alignment of 1RU8 and 2NZ2 (align 1RU8 & i. 11-16, 2NZ2 & i. 11-15) | |||
== alignment of 1RU8 and 2NZ2 (align 1RU8 & i. 11-16, 2NZ2 & i. 11-15)== | |||
[[Image:Dalii.PNG]] | [[Image:Dalii.PNG]] | ||
| Line 43: | Line 48: | ||
[[Image:1ru8 n 2nz2.png]] | [[Image:1ru8 n 2nz2.png]] | ||
Ligand Binding Sites and Surface Clefts | |||
== Ligand Binding Sites and Surface Clefts == | |||
[[Image:PDBsum11.PNG]] | [[Image:PDBsum11.PNG]] | ||
| Line 50: | Line 56: | ||
Surface Topography | == Surface Topography == | ||
[[Image:Topo.PNG]] | [[Image:Topo.PNG]] | ||
Revision as of 12:38, 24 May 2009
Daniel's page in progress
Useful info
Here is some useful paper. check it out Link to Paper: http://pfam.sanger.ac.uk/family?acc=PF00764 Dali: http://ekhidna.biocenter.helsinki.fi/dali_server/results/20090512-0051-3897fbdf5431ad2e808e66dde1070610/index.html#alignment-7 Interpro: http://www.ebi.ac.uk/interpro/DisplayIproEntry?ac=IPR002761
SCOP Work
http://scop.mrc-lmb.cam.ac.uk/scop/data/scop.b.d.cj.c.b.bb.html
Pymol Work
Surface Structure
Secondary Structure and Location of P-loop (ATP binding site)
1RU8 has 2 domain
Electrostatic Surface Potential
alignment of 1RU8 and 2NZ2 (align 1RU8 & i. 11-16, 2NZ2 & i. 11-15)
1RU8 and 2NZ2
Ligand Binding Sites and Surface Clefts