Arylformamidase Function Slide 3: Difference between revisions
Thomasparker (talk | contribs) No edit summary |
Thomasparker (talk | contribs) |
||
| Line 13: | Line 13: | ||
- Screening of DALI result (see Basma) for functional information. | - Screening of DALI result (see Basma) for functional information. | ||
- Most similar | - Most similar structure well-characterised: | ||
''Byun JS, Rhee JK, Kim ND, Yoon J, Kim DU, Koh E, Oh JW, Cho HS.'' | ''Byun JS, Rhee JK, Kim ND, Yoon J, Kim DU, Koh E, Oh JW, Cho HS.'' | ||
'''[http://www.ncbi.nlm.nih.gov/pubmed/17625021?ordinalpos=1&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum Crystal structure of hyperthermophilic esterase EstE1 and the relationship between its dimerization and thermostability properties.]''' | '''[http://www.ncbi.nlm.nih.gov/pubmed/17625021?ordinalpos=1&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum Crystal structure of hyperthermophilic esterase EstE1 and the relationship between its dimerization and thermostability properties.]''' | ||
BMC Struct Biol. 2007 Jul 12;7:47. | BMC Struct Biol. 2007 Jul 12;7:47. | ||
A | - A thermostable carboxylesterase from an uncultured archaeon - isolated in an environmental sample. | ||
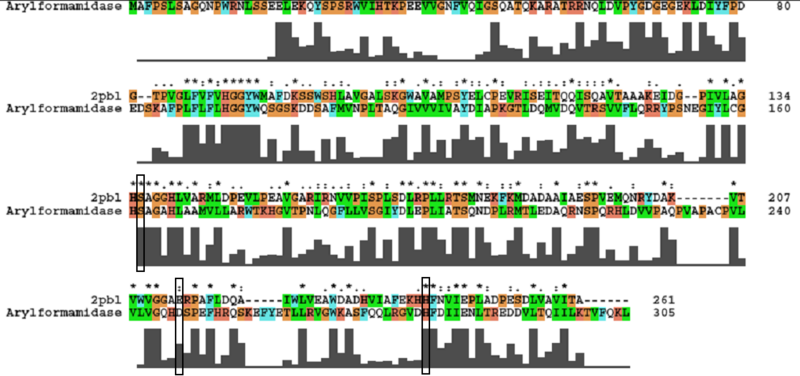
catalytic triad has been identified (how?). To elucidate any functional similarity between 2pbl and 2c7b, conservation of the catalytic triad was assessed (see figure ...). Note: the clustalW alignment was found to differ from the alignment provided as part of the DALI results. All three residues were found to be conserved, though H... and E... were found to match is less conserved regions. Thus, it is possible that such conservation is the result of a chance or poor alignment. This might be explained by the poor sequence similarity between 2c7b and 2pdb (16%). | |||
[[Image:2c7b_alignment.png|centre|framed|'''Conservation of the catalytic triad between 2cb7 and 2pbl.''']] | [[Image:2c7b_alignment.png|centre|framed|'''Conservation of the catalytic triad between 2cb7 and 2pbl.''']] | ||
[[Arylformamidase Function Slide 2| ...Previous slide ]]|[[Arylformamidase| Return to the main page ]]|[[Arylformamidase Function Slide 4| Next slide... ]] | [[Arylformamidase Function Slide 2| ...Previous slide ]]|[[Arylformamidase| Return to the main page ]]|[[Arylformamidase Function Slide 4| Next slide... ]] | ||
Revision as of 12:23, 8 June 2008
Evidence from Similar Sequences
- Catalytic triad identified in paper - Ser162, Asp247, and His279.
- To assess functional similarity, conservation of the catalytic triad was analysed.
- Aspartic acid --> Glutamic acid - Semi-conservative: both polar, acidic.
Evidence from Similar Structures
- Screening of DALI result (see Basma) for functional information.
- Most similar structure well-characterised:
Byun JS, Rhee JK, Kim ND, Yoon J, Kim DU, Koh E, Oh JW, Cho HS. Crystal structure of hyperthermophilic esterase EstE1 and the relationship between its dimerization and thermostability properties. BMC Struct Biol. 2007 Jul 12;7:47.
- A thermostable carboxylesterase from an uncultured archaeon - isolated in an environmental sample.
catalytic triad has been identified (how?). To elucidate any functional similarity between 2pbl and 2c7b, conservation of the catalytic triad was assessed (see figure ...). Note: the clustalW alignment was found to differ from the alignment provided as part of the DALI results. All three residues were found to be conserved, though H... and E... were found to match is less conserved regions. Thus, it is possible that such conservation is the result of a chance or poor alignment. This might be explained by the poor sequence similarity between 2c7b and 2pdb (16%).