Methods 2ece: Difference between revisions
| Line 77: | Line 77: | ||
[[Image:movie0001.png]] | [[Image:movie0001.png]] | ||
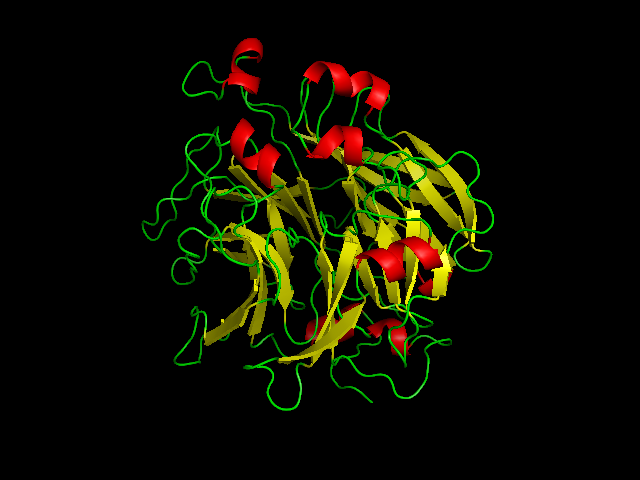
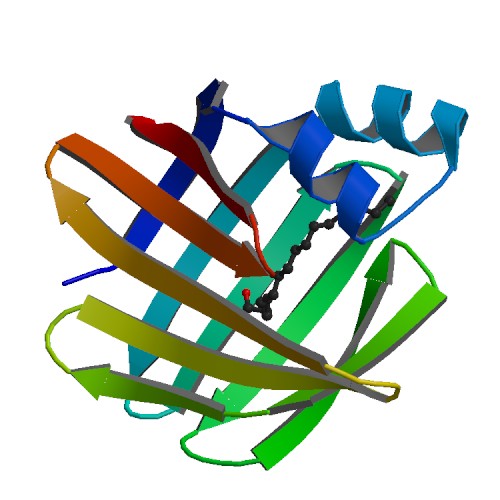
'''Figure 4''' Shows structural helices(red) and beta sheets (yellow) of SBP designed from pymol | |||
Revision as of 04:40, 9 June 2008
STRUCTURAL ANALYSIS
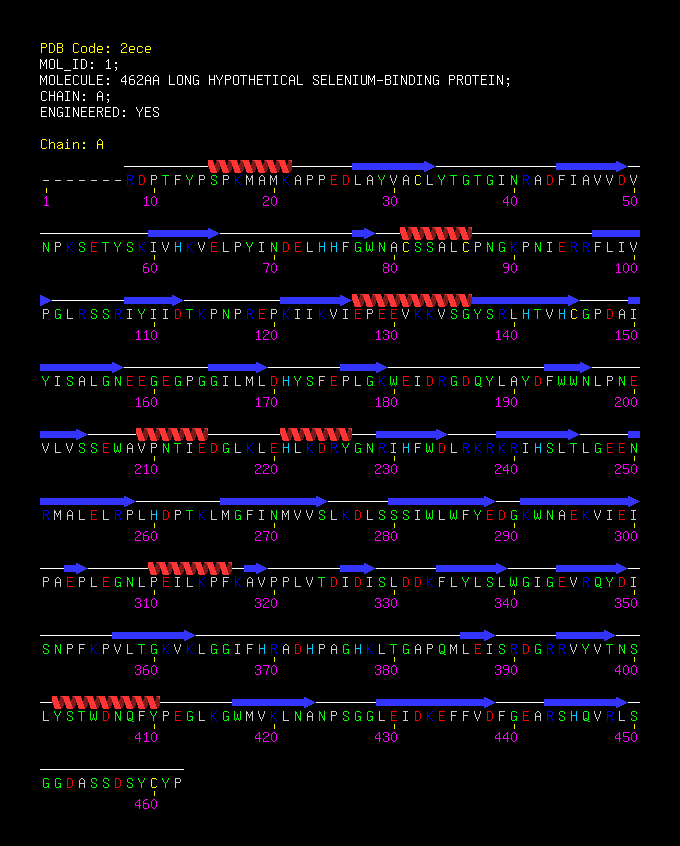
SBP 1 Amino Acid FASTA FORMAT Sequence
>2ECE:A|PDBID|CHAIN|SEQUENCE
MAIVPFKRDPTFYPSPKMAMKAPPEDLAYVACLYTGTGINRADFIAVVDVNPKSETYSKIVHKVELPYINDELHHFGWNA CSSALCPNGKPNIERRFLIVPGLRSSRIYIIDTKPNPREPKIIKVIEPEEVKKVSGYSRLHTVHCGPDAIYISALGNEEG EGPGGILMLDHYSFEPLGKWEIDRGDQYLAYDFWWNLPNEVLVSSEWAVPNTIEDGLKLEHLKDRYGNRIHFWDLRKRKR IHSLTLGEENRMALELRPLHDPTKLMGFINMVVSLKDLSSSIWLWFYEDGKWNAEKVIEIPAEPLEGNLPEILKPFKAVP PLVTDIDISLDDKFLYLSLWGIGEVRQYDISNPFKPVLTGKVKLGGIFHRADHPAGHKLTGAPQMLEISRDGRRVYVTNS LYSTWDNQFYPEGLKGWMVKLNANPSGGLEIDKEFFVDFGEARSHQVRLSGGDASSDSYCYP
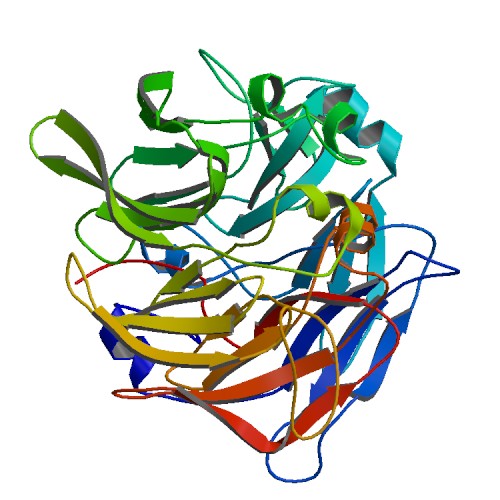
Figure 1 X-ray structure of hypothetical selenium-binding protein from Sulfolobus tokodaii, ST0059 ( http://www.proteopedia.org/wiki/index.php/2ece ) and the JenaLib Jmol viewer showing SBP 1 secondary structure [1]
Query name: SBP 1 ( 2ECE )
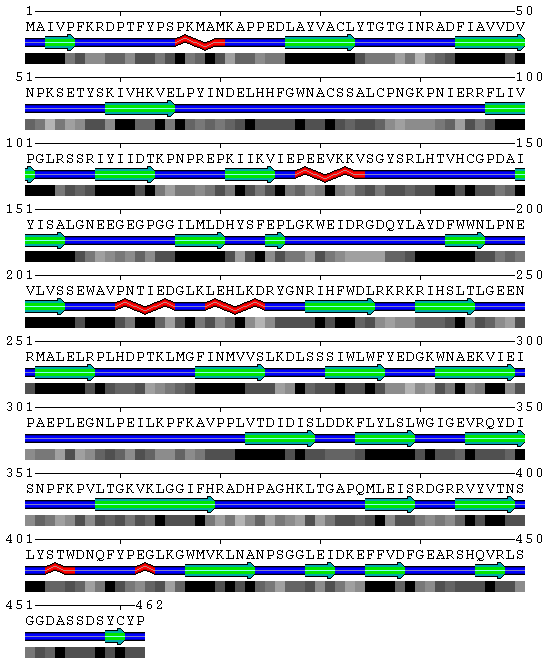
Structure prediction by SABLE
Data source: Derived from the SABLE server prediction
Figure 2 SBP amino acid structure prediction derived from the SABLE server prediction
WARNING! Given sequence appeared to be a soluble protein, no TM domains found!
Output format is the following:
1st line -> residue numeration
2nd line -> query amino acid sequence
3rd line -> trans-membrane domain prediction (T-TM region, N-soluble part)
MAIVPFKRDPTFYPSPKMAMKAPPEDLAYVACLYTGTGINRADFIAVVDVNPKSETYSKI
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
VHKVELPYINDELHHFGWNACSSALCPNGKPNIERRFLIVPGLRSSRIYIIDTKPNPREP
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
KIIKVIEPEEVKKVSGYSRLHTVHCGPDAIYISALGNEEGEGPGGILMLDHYSFEPLGKW
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
EIDRGDQYLAYDFWWNLPNEVLVSSEWAVPNTIEDGLKLEHLKDRYGNRIHFWDLRKRKR
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
IHSLTLGEENRMALELRPLHDPTKLMGFINMVVSLKDLSSSIWLWFYEDGKWNAEKVIEI
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
PAEPLEGNLPEILKPFKAVPPLVTDIDISLDDKFLYLSLWGIGEVRQYDISNPFKPVLTG
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
KVKLGGIFHRADHPAGHKLTGAPQMLEISRDGRRVYVTNSLYSTWDNQFYPEGLKGWMVK
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
LNANPSGGLEIDKEFFVDFGEARSHQVRLSGGDASSDSYCYP
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
Figure 3 SABLE server results.
Figure 4 Shows structural helices(red) and beta sheets (yellow) of SBP designed from pymol
STRUCTURAL COMPARISONS
Surface Structure of SBP: Note the few clefts compared to the DNA isomerase shown below.
Rat fatty acid binding protein 2IFB, a was found to have 92.5% homology to 14 KDa Selenium binding protein purified from rat liver using column chromatography and SDS-Gel techniques.([ref 3])
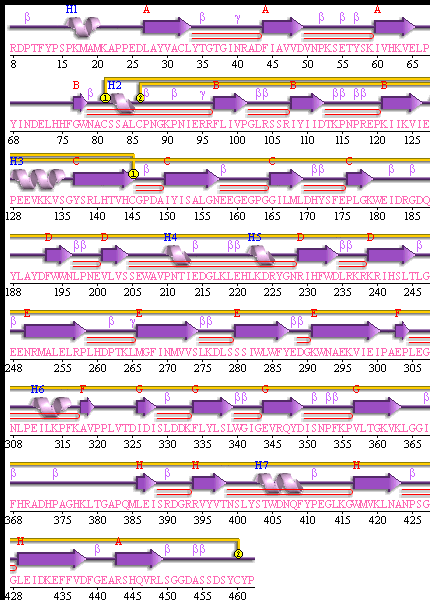
Explore SBP features and structural summary here [3].The domains of SBP are shown here [4] Notice how the domains are similar to the putative Isomerase domains of E.coli below.
1RI6 DOMAINS
2ECE DOMAINS
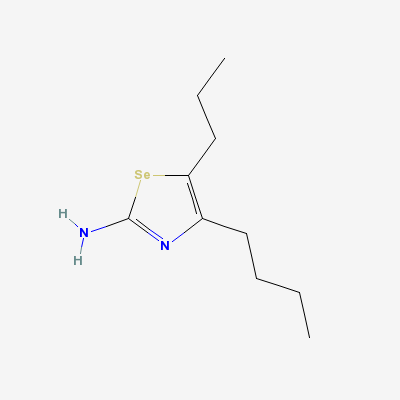
(Complex Of Bovine Odorant Binding Protein (Obp) With A Selenium Containing Odorant)"Image:Ligand of bovine.png" [[5]]
SEQUENCE ANALYSIS
Selenium binding protein 1 (SELENBP1) SELECTED PROTEIN SIMILARITIES Comparison of sequences in UniGene with selected protein reference sequences. The alignments can suggest function of a gene. [6]