Arylformamidase Results
Structure
2PBL Biological Structure
The functional biological structure of arylformamidase is assumed by PDB to be a monomer (see figure 2) even though the 'whole' protein is shown to be interacting with chains A, B, C and D.
2PBL Structural Similarity
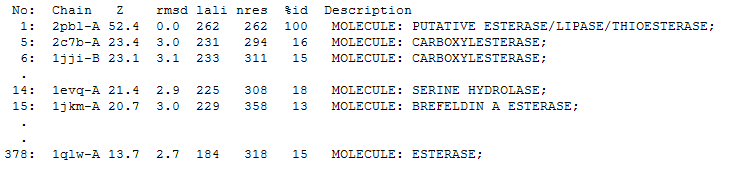
The DALI tool produces proteins that are structurally similar to the protein of interest. The search result showed similarities to be mostly carboxylesterases/hydrolases.
The first significant hit from DALI was a carboxylesterase from a metagenomic Archeaon. The second significant hit was a carboxylesterase of Archaeoglobus fulgidus.
Secondary structure analysis
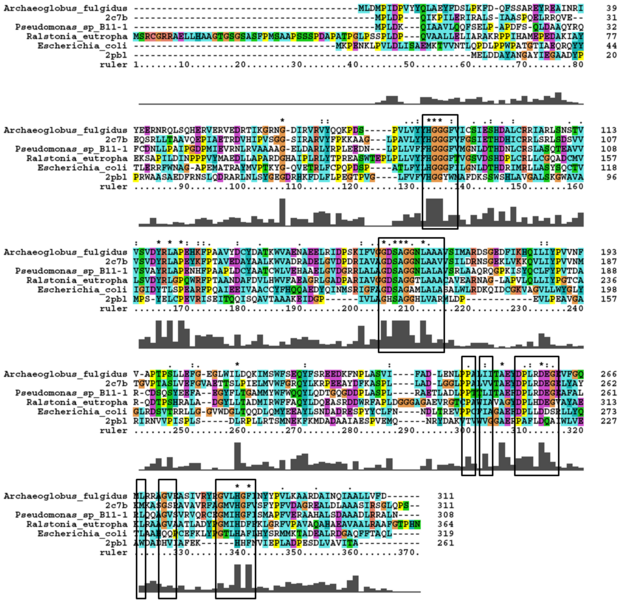
Analysis of arylformamidase's secondary structure with the Archeal carboxylesterases showed the conservation of the order of occurrence of different conformational types. For instance in all three proteins, the first conformation type is a helix and then three beta strands followed by a helix and so on.
The catalytic triad structure
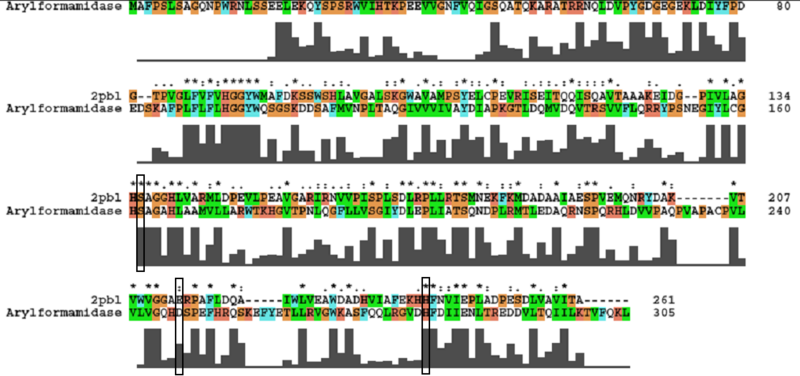
A number of carboxylesterases perform their hydrolysis function using specific catalytic residues. The clustal alignment showed the conservation of the Archeal carboxylesterases catalytic triad in arylformamidase. The residues are Serine (Ser) 137, Glutamate (Glu) 215 and Histidine (His) 242. Residues Ser and His were fully conserved whereas E was semi-conserved. Using the human arylformamidase it was observed that Aspartate (asp) was used for eukaryotes instead of Glu, which is used for prokaryotes.
Other conserved/semi-conserved residues were annotated on the structure of arylformamidase. They are Asp 53, His 69, Gly 70, Gly 71, Tyr 72, Trp 73, Gly 134, Ser 136, Ala 137, Gly 138, His 140, Ser 166, Leu 168, Leu 171, Leu 174, Glu 214, His 241, Val 244 and Leu 248 The blue region in the below structure shows how they all are around the catalytic triad and the unknown ligand. This clearly shows their importance for the function of the protein as they have resisted mutations.
The distance between the catalytic triads can be seen in figure 11. Each of the residues are liked to a turn region. This catalytic triad as stated before is also conserved in the Metagenomic Archea Carboxylesterase (PDB ID 2C7B) and the Archaeoglobus fulgidus Carboxylesterase (PDB ID 1JJI). More so the catalytic triad in Archaeoglobus fulgidus Carboxylesterase is very close to the ligand (see figure 12).
Function
The most similar sequence from the BLAST search with functional information available was an arylformamidase isolated from the liver of Mus musculus (see figure ...). A functional analysis of this arylformamidase has been performed identifying a catalytic triad using site-directed mutagenesis (Pabarcus et al. 2007). Conservation of this catalytic triad with 2pbl was assessed (see figure...). Both residues Ser162 and His279 were found to be identical in relatively conserved regions of the alignment. However, Asp247 had undergone a semi-conservative substitution to glutamic acid. These residues correlated to Ser136, Glu214 and His241 of 2pbl which were subsequently located on the tertiary structure and determined to be sufficiently proximal to one another for catalysis (see figure...).
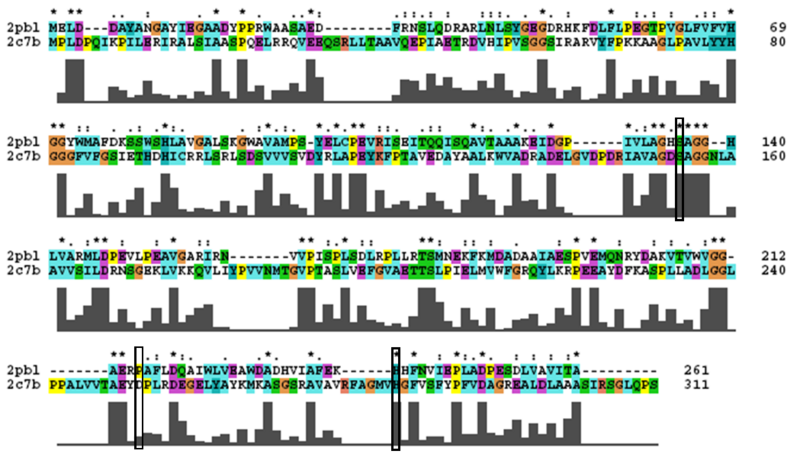
2PBL was found to share most structural similarity with a thermostable carboxylesterase from an uncultured archaeon (PDB ID: 2C7B; see figure ...). 2C7B shares a 16% sequence identity with 2PBL. From its structure, a catalytic triad has been identified with the residues Ser154, Asp251 and His281 (how?)(Byun, et al. 2007). To substantiate any functional similarity between 2PBL and 2C7B, conservation of the 2C7B catalytic triad was analysed (see figure ...). Both Ser154 and His281 matched, but Asp251 was not conserved at all, being replaced for a proline - a nonpolar, neutral amino acid. The sequence surrounding His281 was conserved to a lesser extent than in arylformamidase.
A number of proteins from the hormone-sensitive lipase (HSL) class of lipolytic enzymes as identified by Byun, et al. (2007) was found within top-scoring results of the DALI search (see figure...). To characterise sequence similarity, a ClustalW alignment of the amino acid sequences for these structures was performed (see figure ...).
Sequence & Homology
Figure 1 shows that the query sequence "Arylformamidase" grouped with bacterial sequences, shown cloured in Blue. The bootstrap values reveal low confidence with many of the nodes occurring lower down on the phylogenetic tree revealing a possible explanation for certain closely related species to be grouped into separate clades. Interestingly, despite low bootstrap scores, the grouping reliably separates prokaryotes from eukaryotes and homologous eukaryotes include yeasts and moulds, plants, invertebrates and vertebrates.
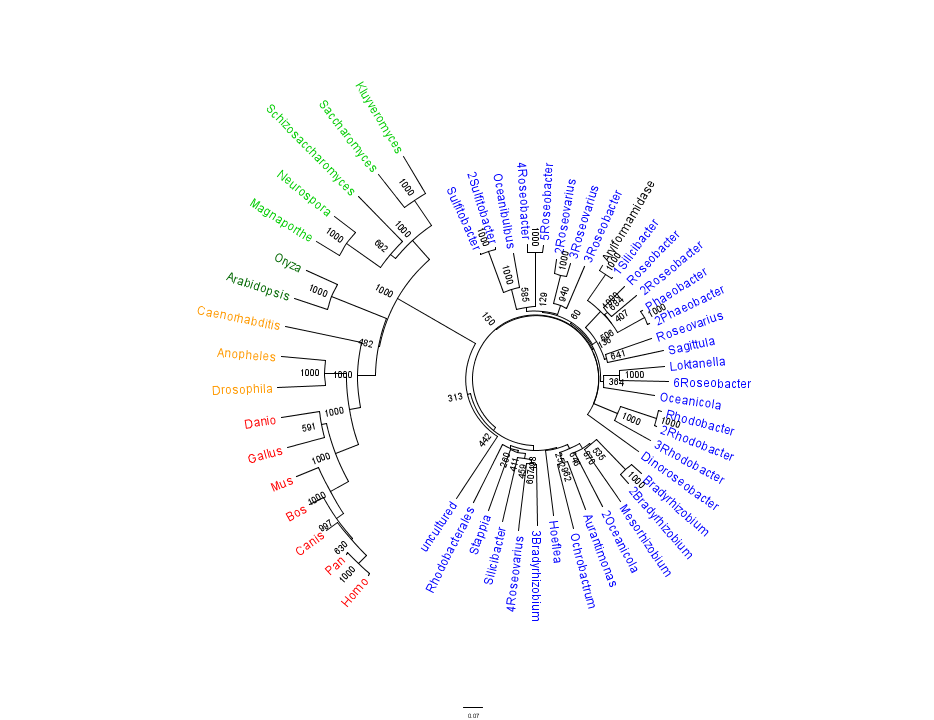
To further elucidate the phylogeny of 2pbl, its human homologue, Arylformamidase, was queried in a BLAST search. The top scoring matches of bacterial homologues, present in Figure 1, were appended with top scoring matches of eukaryotic homologues. The human homologue, Arylformamidase, has a 26.28% sequence similarity. Despite this low score, multiple sequence alignment revealed that key regions were highly conserved between bacterial and eukayotic homologues.
Figure 2 is largely consistent with traditional taxonomic groupings of organisms. Specifically, it reveals greater statistical confidence in the separation of prokaryotes (Blue and Green) and eukaryotes (invertebrates are shown in Orange; vertebrates are in Red).
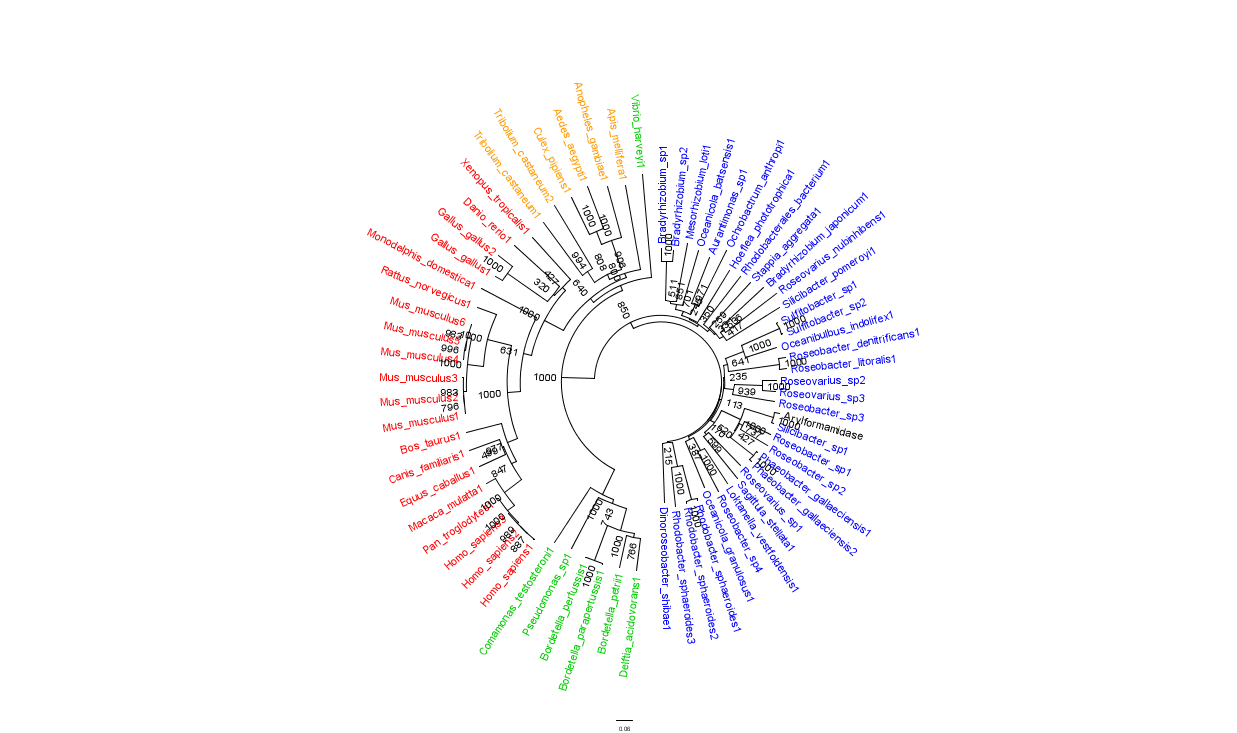
In general, members of the same genus have been grouped together on these phylogenetic trees with some notable exceptions. For instance, Silicibacter, the species from which we derived our protein, occurs on disparate branches of the tree.