Structure ERp18
Methods
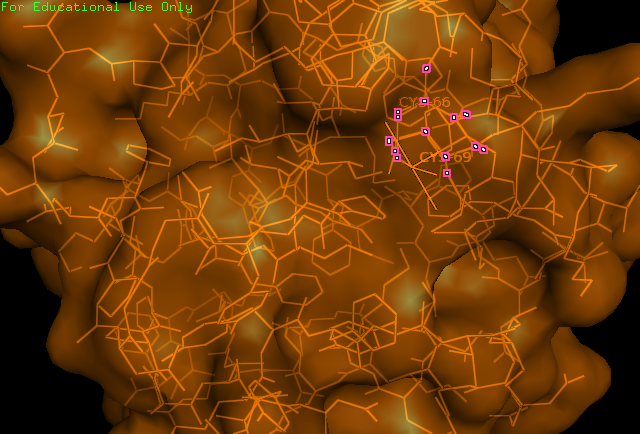
Study of the ERp18 protein (ID = 1sen) began by obtaining its file from the Protein Data Bank (PDB) and visualising the 3D structure of the protein using PyMOL. It was known from study about its function that ERp18 contains a CXXC motif, characteristic of all thiol-disulfide oxidoreductases - this motif was identified as 66-CGAC-69 of the amino acid sequence of mature ERp18. These cysteine residues were found to be localised to the surface of the protein and were predicted to form the catalytic site. Next, conserved sequences identified by a ClustalW multiple alignment from evolution study were highlighted on the protein. Following this, searches were made with PDBsum, SCOP, and Prosite for more information about the folding and domain-annotation of the protein. Secondary structures were provided by PDBsum. After completing these fundamental protein structure searches, published papers were consulted and their findings on ERp18 were compared with our findings.
Results
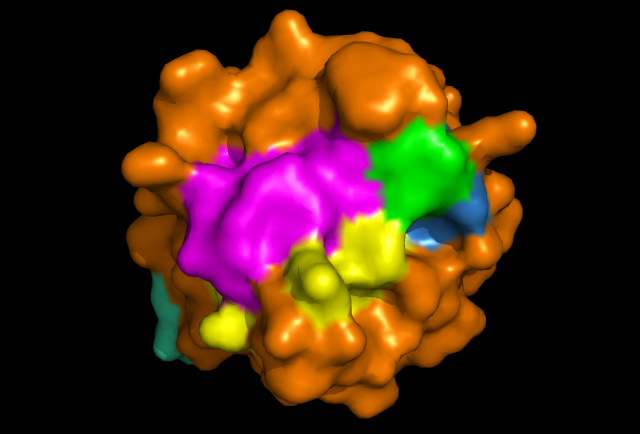
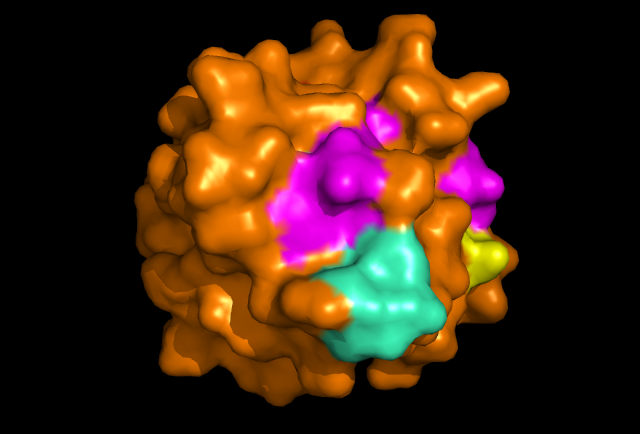
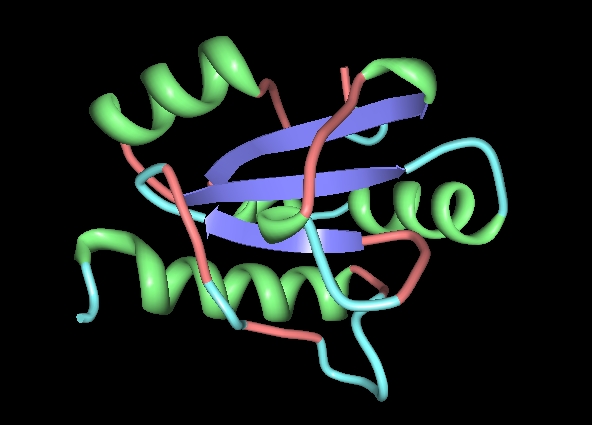
[[Image: Image_of_ERp18_with_conserved_residues.png|framed|Figure 4: PyMOL image displaying the conserved sections of a ribbon diagram of ERp18. Notice the conservation of the three core beta sheets - probably vital for maintaining stability and correct folding of the protein, as well as correct catalytic activity.
Dali Summary
No: Chain Z rmsd lali nres %id PDB Description 1: 1sen-A 31.6 0.0 134 134 100 PDB MOLECULE: THIOREDOXIN-LIKE PROTEIN P19; 2: 3f9u-A 13.0 2.2 114 145 12 PDB MOLECULE: PUTATIVE EXPORTED CYTOCHROME C BIOGENESIS- 3: 3f9u-B 12.3 2.2 114 148 12 PDB MOLECULE: PUTATIVE EXPORTED CYTOCHROME C BIOGENESIS- 4: 2fwe-A 11.7 2.4 107 122 21 PDB MOLECULE: THIOL:DISULFIDE INTERCHANGE PROTEIN DSBD; 5: 2fwf-A 11.6 2.4 107 123 21 PDB MOLECULE: THIOL:DISULFIDE INTERCHANGE PROTEIN DSBD; 6: 1se1-C 11.1 2.1 103 231 22 PDB MOLECULE: THIOL:DISULFIDE INTERCHANGE PROTEIN DSBD; 7: 1vrs-F 11.1 2.1 103 118 22 PDB MOLECULE: THIOL:DISULFIDE INTERCHANGE PROTEIN DSBD; 8: 1vrs-D 11.1 2.2 102 118 22 PDB MOLECULE: THIOL:DISULFIDE INTERCHANGE PROTEIN DSBD; 9: 1se1-A 11.1 2.2 102 239 22 PDB MOLECULE: THIOL:DISULFIDE INTERCHANGE PROTEIN DSBD; 10: 1ep7-B 10.9 2.4 102 112 13 PDB MOLECULE: THIOREDOXIN CH1, H-TYPE; 11: 2fwg-A 10.9 2.7 106 122 22 PDB MOLECULE: THIOL:DISULFIDE INTERCHANGE PROTEIN DSBD; 12: 2fwh-A 10.9 2.2 102 117 23 PDB MOLECULE: THIOL:DISULFIDE INTERCHANGE PROTEIN DSBD; 13: 1uc7-A 10.8 2.2 104 124 22 PDB MOLECULE: THIOL:DISULFIDE INTERCHANGE PROTEIN DSBD; 14: 1ep8-B 10.8 2.5 103 112 13 PDB MOLECULE: THIOREDOXIN CH1, H-TYPE; 15: 1ep8-A 10.7 2.5 103 112 13 PDB MOLECULE: THIOREDOXIN CH1, H-TYPE; 16: 2ju5-A 10.6 2.2 105 144 17 PDB MOLECULE: THIOREDOXIN DISULFIDE ISOMERASE; 17: 1ep7-A 10.6 2.6 103 112 13 PDB MOLECULE: THIOREDOXIN CH1, H-TYPE; 18: 1vrs-E 10.6 2.2 102 118 22 PDB MOLECULE: THIOL:DISULFIDE INTERCHANGE PROTEIN DSBD; 19: 1uc7-B 10.5 2.3 104 124 23 PDB MOLECULE: THIOL:DISULFIDE INTERCHANGE PROTEIN DSBD; 20: 3d22-A 10.4 2.8 108 129 13 PDB MOLECULE: THIOREDOXIN H-TYPE; 21: 3d21-A 10.4 2.5 101 111 16 PDB MOLECULE: THIOREDOXIN H-TYPE; 22: 3d21-B 10.3 2.5 101 111 16 PDB MOLECULE: THIOREDOXIN H-TYPE; 23: 3fk8-A 10.2 2.6 106 131 17 PDB MOLECULE: DISULPHIDE ISOMERASE; 24: 2vlv-B 10.2 2.3 99 113 22 PDB MOLECULE: THIOREDOXIN H ISOFORM 2.; 25: 1se1-B 10.2 2.2 102 243 22 PDB MOLECULE: THIOL:DISULFIDE INTERCHANGE PROTEIN DSBD; 26: 2vlu-B 10.2 2.3 100 112 22 PDB MOLECULE: THIOREDOXIN H ISOFORM 2.; 27: 2vm1-A 10.2 2.3 99 110 16 PDB MOLECULE: THIOREDOXIN H ISOFORM 1.; 28: 2vlv-A 10.1 2.4 100 111 22 PDB MOLECULE: THIOREDOXIN H ISOFORM 2.; 29: 2vm2-A 10.1 2.3 99 109 16 PDB MOLECULE: THIOREDOXIN H ISOFORM 1.; 30: 2vlt-B 10.1 2.4 100 110 22 PDB MOLECULE: THIOREDOXIN H ISOFORM 2.;
Figure 5: The top thirty hits from a DALI search with 1sen-A. DALI lists proteins in order of their structural similarity. The protein names displayed are almost entirely thiol:disulfide interchange proteins and thioredoxins (thioredoxins being a related protein to the catalytic domain of thiol:disulfide interchange proteins), with the exception of 'putative exported cytochrome c biogenesis-related protein' for 2 and 3. However, hits 2 and 3 contain the CXXC motif and are therefore likely to be members of the thioredoxin family. Interestingly, the percent identification (%id) of these hits ranges between 12-23% - quite low considering the degree of structural homology as assigned by the rmsd value, for which structure similarity is considered significant if the value is less than 3.5.
PDBsum
PDB id: 1sen Name: Structural genomics, unknown function Title: Endoplasmic reticulum protein rp19 o95881
Structure: Thioredoxin-like protein p19. Chain: a. Synonym: endoplasmic reticulum protein erp19. Engineered: yes
Source: Homo sapiens. Human. Organism_taxid: 9606. Gene: tlp19. Expressed in: escherichia coli. Expression_system_taxid: 562. Other_details: the protein was cloned, expressed and purified by the secsg human protein production group (t.A. Dailey, m. Mayer) under the direction of h.A. Dailey.
UniProt: O95881 (TXD12_HUMAN) SAS Seq: 172 a.a. Struc: 135 a.a. Key: PfamB domain Secondary structure CATH domain
Enzyme class: E.C.1.8.4.2 [IntEnz] [ExPASy] [KEGG] [BRENDA]
Reaction: 2 glutathione + protein-disulfide = glutathione disulfide + protein- dithiol (see diagram below)
Resolution: 1.20Å
R-factor: 0.162
R-free: 0.183
Authors: Z.-J.Liu,L.Chen,W.Tempel,A.Shah,D.Lee,T.A.Dailey,M.R.Mayer, J.P.Rose,D.C.Richardson,J.S.Richardson,H.A.Dailey,B.-C.Wang Southeast Collaboratory For Structural Genomics (Secsg)
Key ref: z.-j.liu et al. Endoplasmic reticulum protein Rp19. To be Published, xsi:nil="true" />.
Date: 17-Feb-04
Release date: 13-Jul-04
Related entries: O95881 related db: targetdb
SCOP Summary
Protein: Thioredoxin-like protein p19, TLP19 from Human (Homo sapiens) [TaxId: 9606] Lineage:
1. Root: scop
2. Class: Alpha and beta proteins (a/b) [51349]
Mainly parallel beta sheets (beta-alpha-beta units)
3. Fold: Thioredoxin fold [52832]
core: 3 layers, a/b/a; mixed beta-sheet of 4 strands, order 4312; strand 3 is antiparallel to the rest
4. Superfamily: Thioredoxin-like [52833]
5. Family: Thioltransferase [52834]
6. Protein: Thioredoxin-like protein p19, TLP19 [110604]
7. Species: Human (Homo sapiens) [TaxId: 9606] [110605]
Discussion
The localisation of the CXXC motif to a highly conserved surface region indicates that it forms the active site. Conservation of the internal B-sheets is probably vital for the correct conformation of the protein. Interestingly, second surface region of high homology was identified and it could be the site of interaction with another protein, or possibly another copy of itself. The literature identifies ERp18 as a homodimer, with a single protein having a molecular weight of 16.4 kDa. Structure determines function etc
References
1. Jeong, W., Lee, D., Park, S., & Rhee, S.G., (2008) "ERp16, an Endoplasmic Reticulum-resident Thiol-disulfide Oxidoreductase" THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 283, NO. 37, pp. 25557–25566.
2. Liu, F., Rong, Y., Zeng, L., Zhang, X., & Han, Z.,(2003) "Isolation and characterization of a novel human thioredoxin-like gene hTLP19 encoding a secretory protein" Gene, Volume:315, pp. 71–78.
3. Alanen, H. I. et al. (2003) "Functional Characterization of ERp18, a New Endoplasmic Reticulum-located Thioredoxin Superfamily Member" Vol. 278, No. 31, Issue of August 1, pp. 28912–28920.