C1orf41 Results
Structure
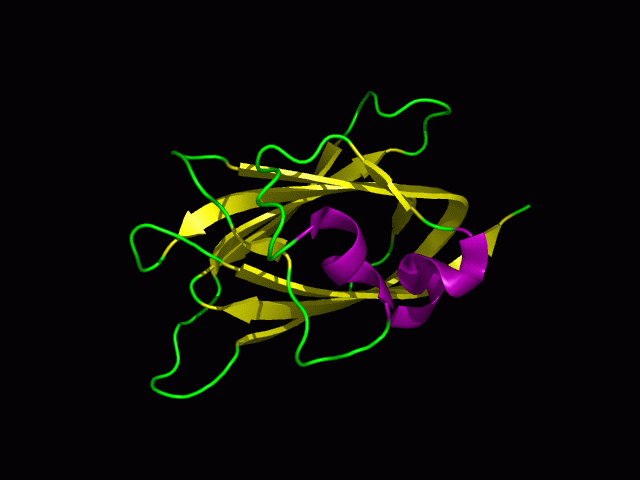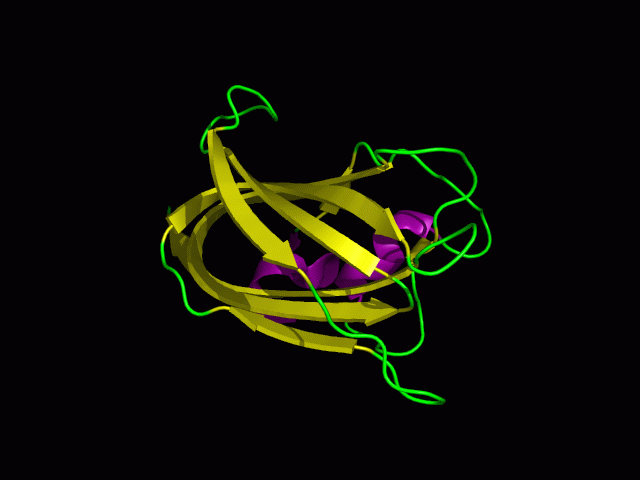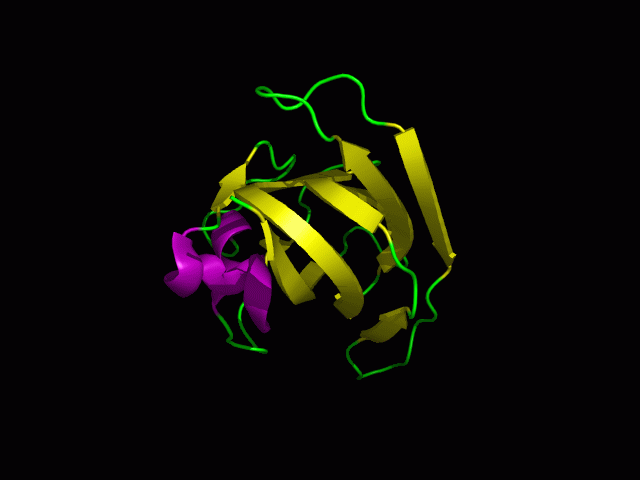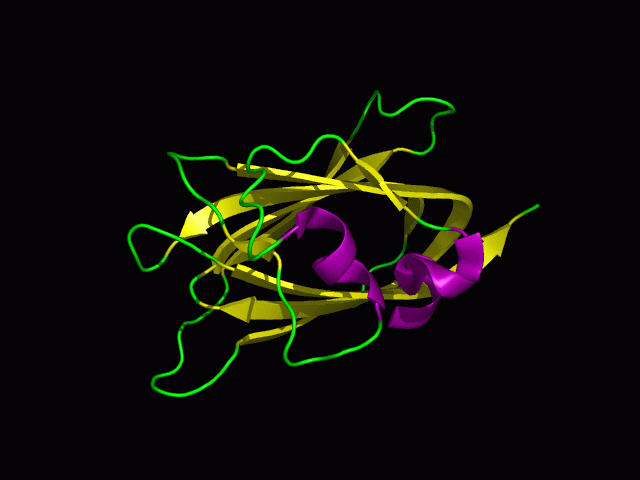
Protein Structure
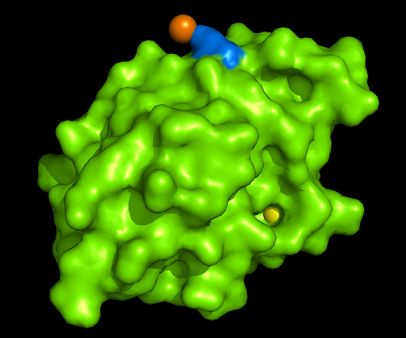
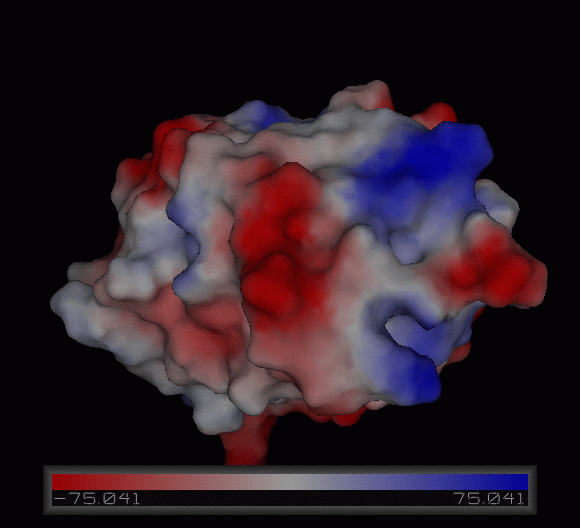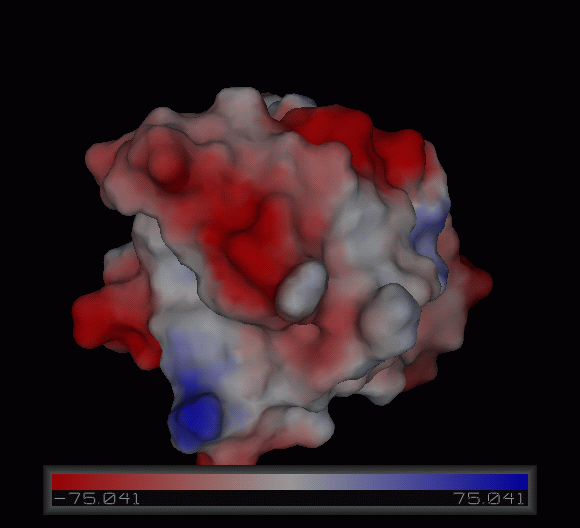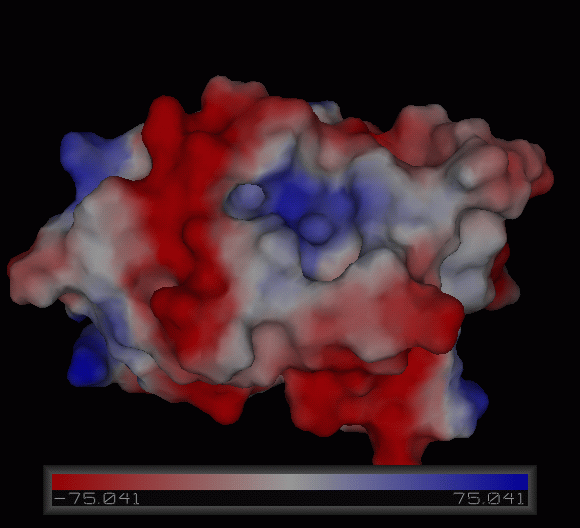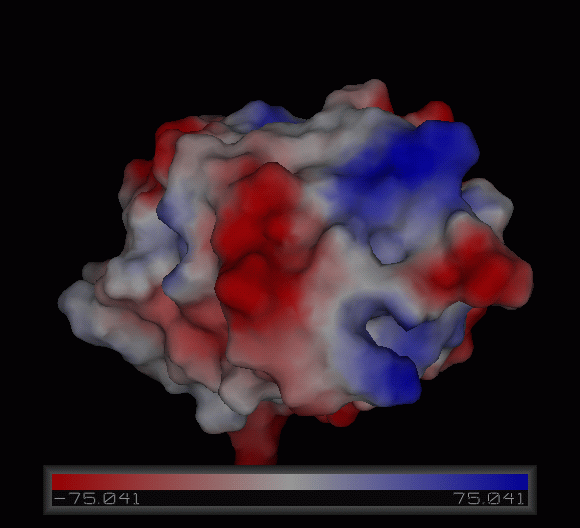
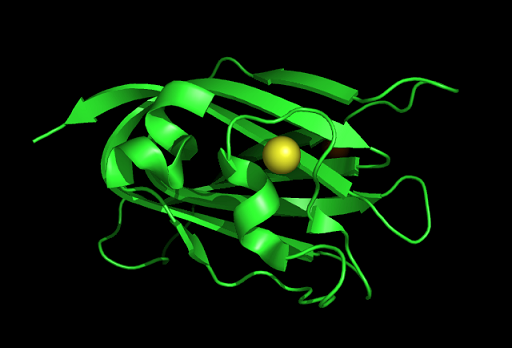
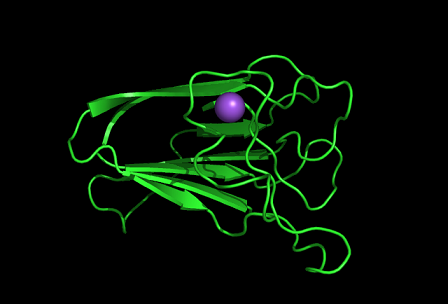
PDB ID: 1TVG Information from the PDB stated that x-ray diffraction was ueed to solve the structure of this protein with an R value of 0.215 and at 1.6Å. Two ligands were present in the crystal structure, a calcium (II) ion and a samarium (III) ion. This protein has 153 residues and its secondary structure consists of two helices and nine beta sheet strands.
Scop result
Tertiary structure
Structural analysis
1)Structural similarities
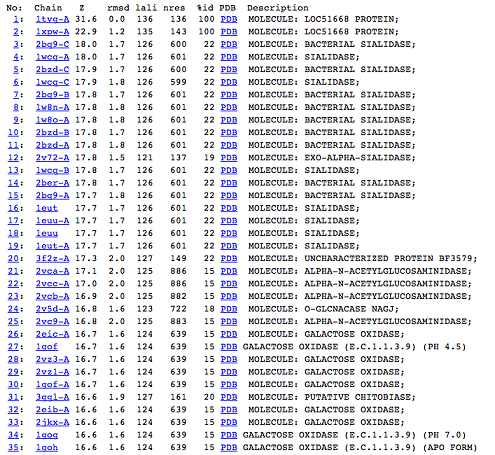
DALI was used to find proteins that are similar in structure to c1orf41. The results showed that sialidases, alpha-N-acetylglucosaminidases and galactose oxidases have similar structures to this protein.
2)Domain Classification
3)Possible ligand binding sites
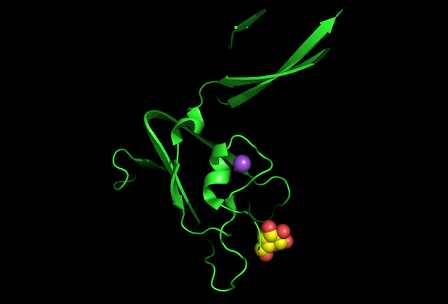
Observations and comparisons of several proteins from the DALI result with our protein showed that there is a similar position on each proteins where a metal ion is located.
Figure 10: Pymol figure displaying possible metal binding position on c1orf41. Orange is the residues that may be involved in coordinating the metal and yellow is the calcium ion. The residues are based on CastP result.
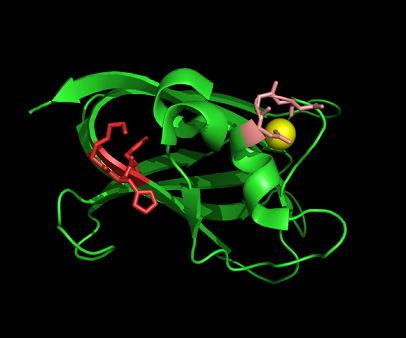
Nest analysis by Profunc suggested two other ligand binding sites. Nests are structural motifs that commonly found in functionally important regions of protein structures.
Figure 11: Nest 1 and has scores higher than 2. So, they are more likely to be functional. His54 and Lys55 NH atoms are accessible from a large surface cleft of the protein. The cleft is also deep indicating that the nest is functionally important. The residues of Nest 2 are part of the loop that coordinates the Ca (II)ion.
Figure 12: Pymol representation of Nest 1 and 2.
Figure 13: Arrows indicating the pockets of the nests that may be accessible to ligands.
Evolution
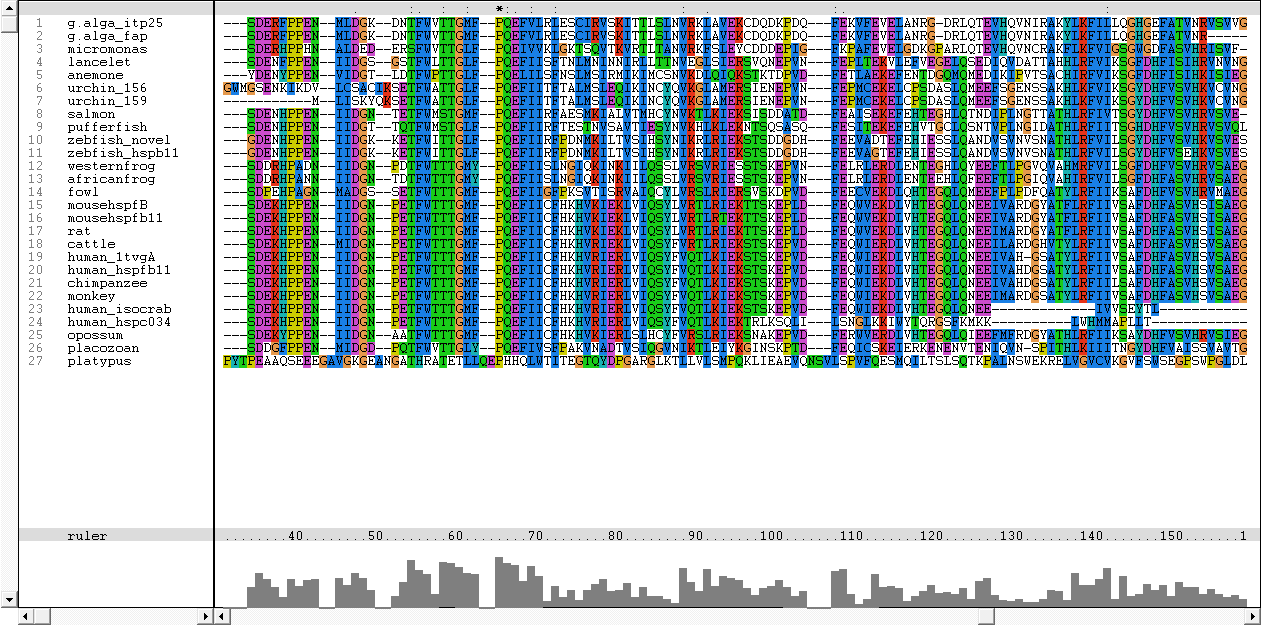
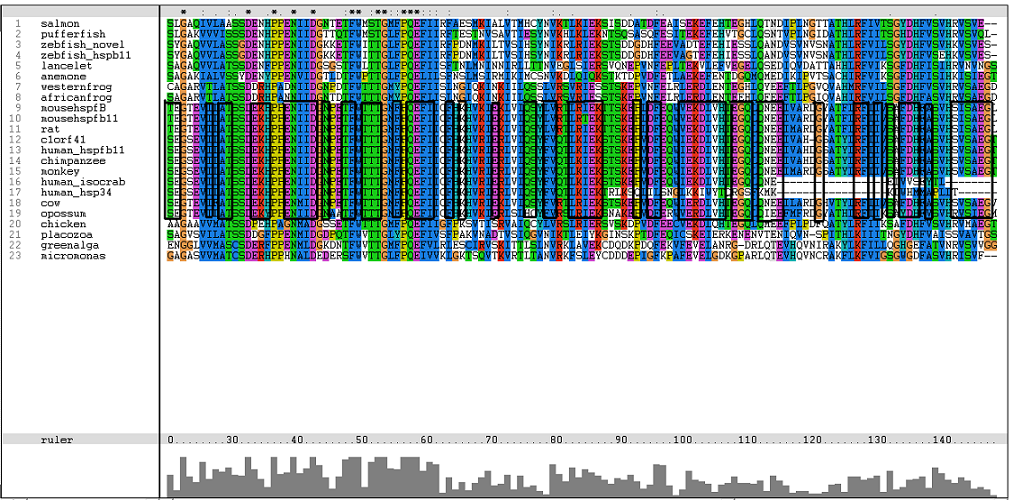
Results from Blast showed that c1orf41 have high homology to small heat shock proteins which gave us a hint that the protein may be a heat shock protein (Figure 14). However, initial multiple sequence alignment showed many gaps and few conserved regions (Figure 15).
The sequences from platypus, urchin_156, urchin_159 and g.alga_fap were excluded from further investigation. This was due to poor alignment of platypus, urchin_156 and urchin_159 with the other organisms. In addition, the sequence for g.alga_fap was removed as it was a truncated sequence of g.alga_itp25.
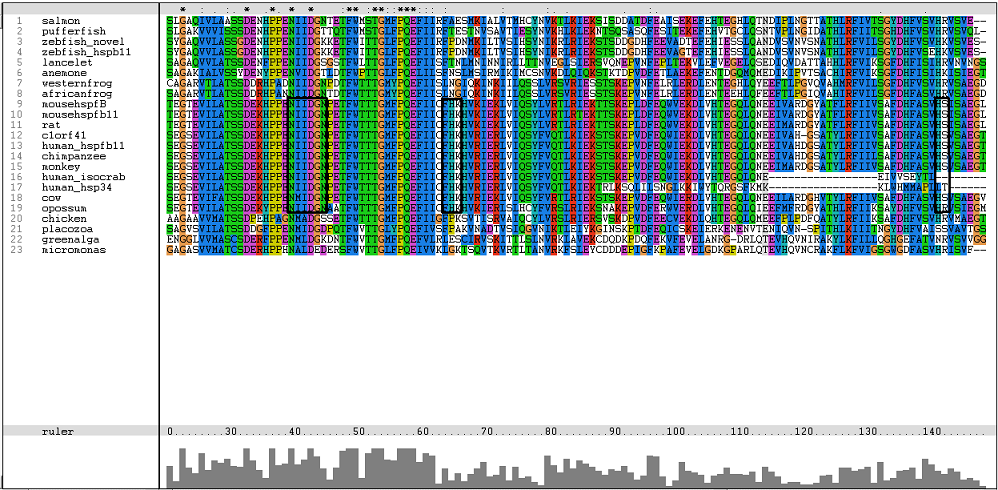
One of the characteristic of a small heat shock protein is that they have a conserved alpha B crystallin domain which can also be observed in c1orf41 as shown in figure 16 below:
The conserved residues found in c1orf41 and its homologues fall in the alpha B crystallin domain. But more conserved regions are observed in higher terrestrial eukaryotes. With this, we are more confident that c1orf41 is a heat shock protein.
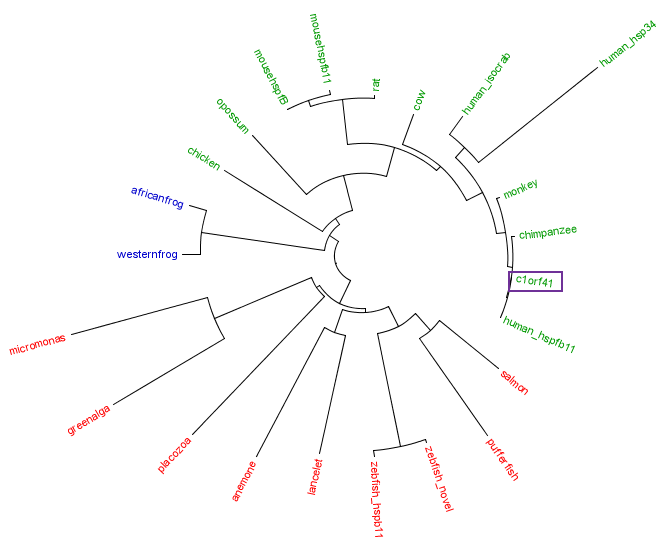
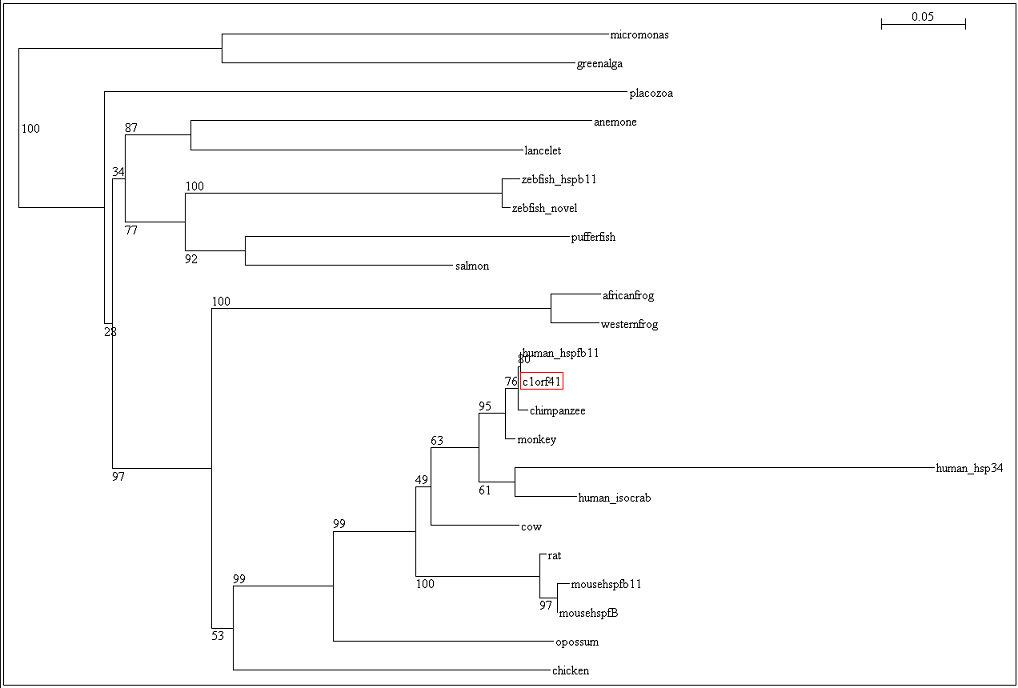
The residues of the possible binding site were highlighted in black in figure 17 below. However, these residues were only conserved in terrestrial eukaryotes. During evolution, the protein has evolve and adopt different function in marine eukaryotes, amphibian and terrestrial eukaryotes. The evolutionary relationship of this protein is shown in figure 18.
To ensure that the reliability of the evolutionary relationship of c1orf41 and its homologues, the tree was bootstrap with 100 replicates (Figure 19). Bootstrap value of more than 75 indicated that we are confident in the branch whereas bootstrap value of less than 50 indicated no confidence in the branch over an alternative