Talk:Protein Function
Information From 8th May 2007
MIF4G is the middle domain of eukaryotic initiation factor 4G (eIF4G). It also occurs in NMD2p (non-sense mediated mRNA decay protein - it is involved in the non-sense mediated decay of mRNAs containing premature stop codons) and in CBP80 (Cap binding protein).
The protein binds eIF4A, eIF3, RNA and DNA Therefore part of function is to bind to RNA
Possibly located in the cytoplasm - See link to LOCATE. Mouse protein of similar seuqence in this location.
MIF4G starts residue 28 Ends 240 (mouse)
It is soluble and non-secreted.
PA74324.2 Riken cDNA 2310075612 Rik Protein - AAH26740, AAH55812(mouse), AAH33759(human)
AAH55812 - Rik Protein Mouse. Present in the cerebellum, Striatum, Eye, Wholebrain, Liver, Hippocampus, Hematopoietic Stem Cells and Kidney Accession No: BC055812.1
Performed a MultiLoc prediction that determines location of the protein based on Amino Acid sequence and the presence etc of a N-termial targeting sequence. There is a 0.93 Probability that the protein is cytoplasmic. Now I have to find specific location, what the protein binds to and the structure of what it binds to. If i can identify the structure of the binding domain then I can predict to some extent the structure or a very small piece of the structure ie active site and can use this to perform function based analysis?
ProFunc Analysis:
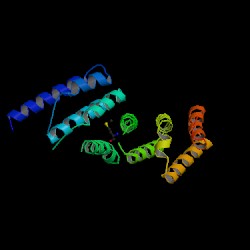
Showed that the domain contains an ARM repeat. Further research into this will be done. Eliza found the same thing.
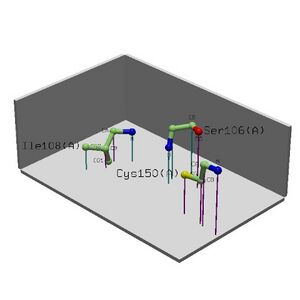
This shows that there are many binding sites. To get to this image follow the link under the Cleft Sites analysis on the ProFunc results page.
Still need to ID what is the significance of all the results uncovered by ProKnow
Will go into this more next week
But it is interesting to know for the time being that both eliza and I have found that the function has something to do with the methylated cap on RNA and that it is this process with-in the cytoplasm (as opposed to in the nucleus).
Site: Showed that Danio Renio is 99.5% likely to be a match in structure to 2i2O> we can then make an inference that since they are both in the same region (Double Check this on Locate) and they have the same structure and x % similarity in sequence then it is likely they are related in function.
Same alignment results for 1hu3 eIF4Gii. http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl?pdbcode=ay65&pdb_type=PROFUNC&code=075103&template=sitehit.html&profunc=TRUE&u=&l=2.1&o=SITE
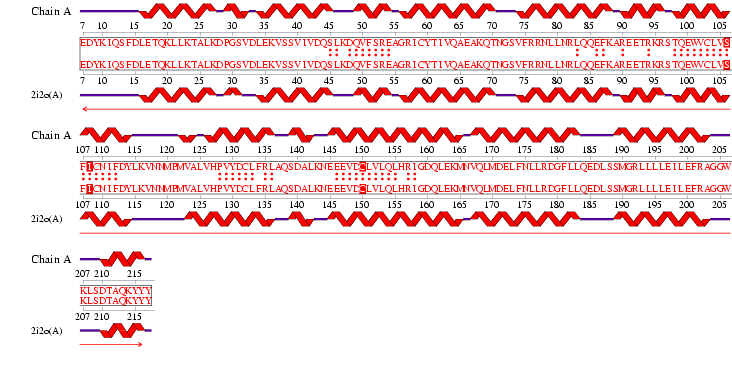
NEST Analysis: Found 3 nests within Structure. This provided possible functional residues.
Figure 1.0: Alignment obtained from ProFunc NEST
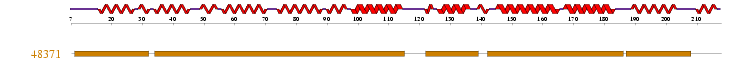
Superfamily Results showed 1 sequence motif found in the sequence provided. http://supfam.org/SUPERFAMILY/cgi-bin/scop.cgi?sunid=48371 This revealed the presence of ARM Repeats
Figure 1.1: Superfamily analysis revealed 1 sequence motif in the sequence.
ProKnow Analysis
Table shows that the most likely molecular function for our protein is RNA binding this is infered by genetic interaction. Most of the Biological processes ID'd are from a traceable author statement. Number of clues are 6 and 4 respectively. 1-2 is considered weak therefore 4 and 6 probably arent greatly significant but perhaps high enough to make some inferrences.
When looking at the Master Table from results - note the following:
*Clue 1 Frequency of the ontolgies obtained from Blast hits
*Clue 2 Score for the ontology from Blast Evalues. The best evalue available for the ontology is taken (only 4
digits after decimal is shown).
*Clue 3 Frequency of ontologies from 3D motifs
*Clue 4 Score of ontologies from 3D motifs based on conservation. It is the average of scores from the motifs
associated with the ontology.
*Clue 5 Score of ontologies from 3D folds. The best Z_score available for the function is taken.
*Clue 6 Frequency of ontologies from 3D folds
*Clue 7 Frequency of ontologies from DIP search
*Clue 8 Score of ontologies from PROSITE search based on conservation. It is the average of scores from the motifs
associated with the ontology.
*Clue 9 Frequency of ontologies from PROSITE search
*Clue 10 Frequency of ontologies from PROLINKS search
Our results had a very high reading in clue 8. Does this mean that the sequ is highly conserved??
Article about eIF4GIII Protein - http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=11172724
Most similar structurally to zebra fish - shows that the sequences within the domain are not similar. The structures among the different proteins of the domain are similar but the sequences are different. Does this make all the phylogeny and sequ analysis stuff kind of redundant??
PDB Search on Danio Renio 1hu3
http://www.rcsb.org/pdb/explore/explore.do?structureId=1HU3
Papers on RNA/DNA Binding Proteins
http://www.wormbook.org/chapters/www_RNAbindingproteins/RNAbindingproteins.html
http://www.molecular-cancer.com/content/3/1/24
Obtained Sequences
Human - Protein Sequ
mgepsreeyk iqsfdaetqq llktalkvac fetedgeysv cqrsysncsr lmpsrcntqy
rdpgavdlek vanvivdhsl qdcvfskeag rmcyaiiqae skqagqsvfr rgllnrlqqe
yqareqlrar slqgwvcyvt ficnifdylr vnnmpmmalv npvydclfrl aqpdslskee
evdclvlqlh rvgeqlekmn gqrmdelfvl irdgfllptg lsslaqllll eiiefraagw
kttpaahkyy ysevsd
>AAH26740 ARM repeat, position: 13-208 (Mouse)
SFDAQTQQLLKTALKDPGAVDLERVANVIVDHSLQDCVFSKEAGRMCYAIIQAESKQAGQSVFRRGLLNRLQKEYDAREQ
LRACSLQGWVCYVTFICNIFDYLRVNNMPMMALVNPVYDCLFQLAQPESLSREEEVDCLVLQLHRVGEQLEKMNGQRMDE
LFILIRDGFLLPTDLSSLARLLLLEMIEFRAAGWK
Mouse - Protein
mseasrddyk iqsfdaetqq llktalkdps avdlervanv ivdhslqdcv fskeagrmcy
aiiqaeskqa gqsvfrrgll nrlqkeydar eqlracslqg wvcyvtficn ifdylrvnnm
pmmalvnpvy dclfqlaqpe slsreeevdc lvlqlhrvge qlekmngqrm delfilirdg
fllptdlssl arllllemie fraagwkttp aahkyyysev sd
FASTA - Human
>gi|21707112|gb|AAH33759.1| MIF4G domain containing [Homo sapiens]
MGEPSREEYKIQSFDAETQQLLKTALKVACFETEDGEYSVCQRSYSNCSRLMPSRCNTQYRDPGAVDLEK
VANVIVDHSLQDCVFSKEAGRMCYAIIQAESKQAGQSVFRRGLLNRLQQEYQAREQLRARSLQGWVCYVT
FICNIFDYLRVNNMPMMALVNPVYDCLFRLAQPDSLSKEEEVDCLVLQLHRVGEQLEKMNGQRMDELFVL
IRDGFLLPTGLSSLAQLLLLEIIEFRAAGWKTTPAAHKYYYSEVSD