Function
The predicted function based on the evolution and structure
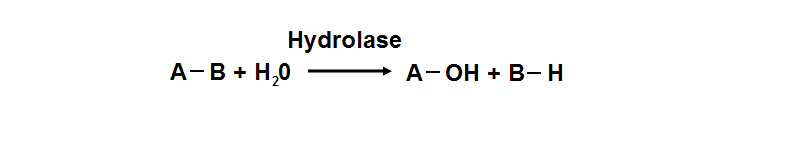
Hydrolyase catalyze the hydrosis of the chemical bond between A and B, resulting of 2 simple molecules
- Hydrolase
- Catalyze hydrolysis reaction
- Addition of the hydrogen and hydroxyl ions of water
- Splitting into 2 or more simpler molecules
- EC class 3
Pfam
|
Hydrolase: domain 1 of 1, from 18 to 224: score 96.2, E = 1e-25 *->ikavvFDkDGTLtdgkeppiaeaiveaaaelgl.........lplee
++av+FD+D+TL+d+ + + ++ + e+ ++l + + +++ ++ +
query 18 VRAVFFDLDNTLIDT-AGASRRGMLEVIKLLQSkyhykeeaeIICDK 63
vekllgrgl.g.erilleggltaell...................d.evl
v l +++ ++ ++ t ++ + +++++ ++++ ++ ++
query 64 VQVKLSKECfHpYSTCITDVRTSHWEeaiqetkggadnrklaeecYfLWK 113
glial.dklypgarealkaLkrrGikvailTggdr.naeallealgla.l
++ ++ l +++++ l +L++ +++ +lT+gdr++++++ ea+++ ++
query 114 STRLQhMILADDVKAMLTELRKE-VRLLLLTNGDRqTQREKIEACACQsY 162
fdviidsdevggvgpivvgKPkpeifllalerlgvkpeevgpevlmVGDg
fd+i++++e + KP+p if + ++ lgv+p ++ +mVGD+
query 163 FDAIVIGGEQK------EEKPAPSIFYHCCDLLGVQPGDC----VMVGDT 202
vnDapalaa.AGv.gvamgngg<-*
+ +++ + +AG+++++++n +
query 203 LETDIQGGLnAGLkATVWINKS 224
|
The alignments of the top-scoring domains of 2gfh (query) protein using Pfam 21.0 (Janelia Farm). (http://pfam.janelia.org)
- Shows the highest scored match - Hydrolase
- Score = 96.2
- E value = 1e-25
Gene Ontology
List of all matched protein name terms for 2gfh. The score in red is a measure of how strongly the term is predicted from the hits obtained by
the different methods. The scores in blue show each method’s contribution to the total score (with the number of relevant
sequences/structures shown in brackets in grey).(http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/)
- Higest score in GO - Hydrolase
- Followed by - Phosphatase
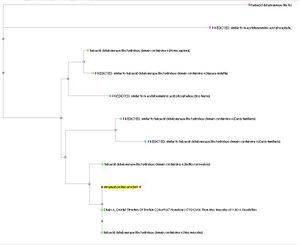
- Polymer: Haloacid dehalogenase-like hydrolase domain containing 4
- Molecular Function: None
- Biological Process: None
- Cellular Component: None
Surface Properties
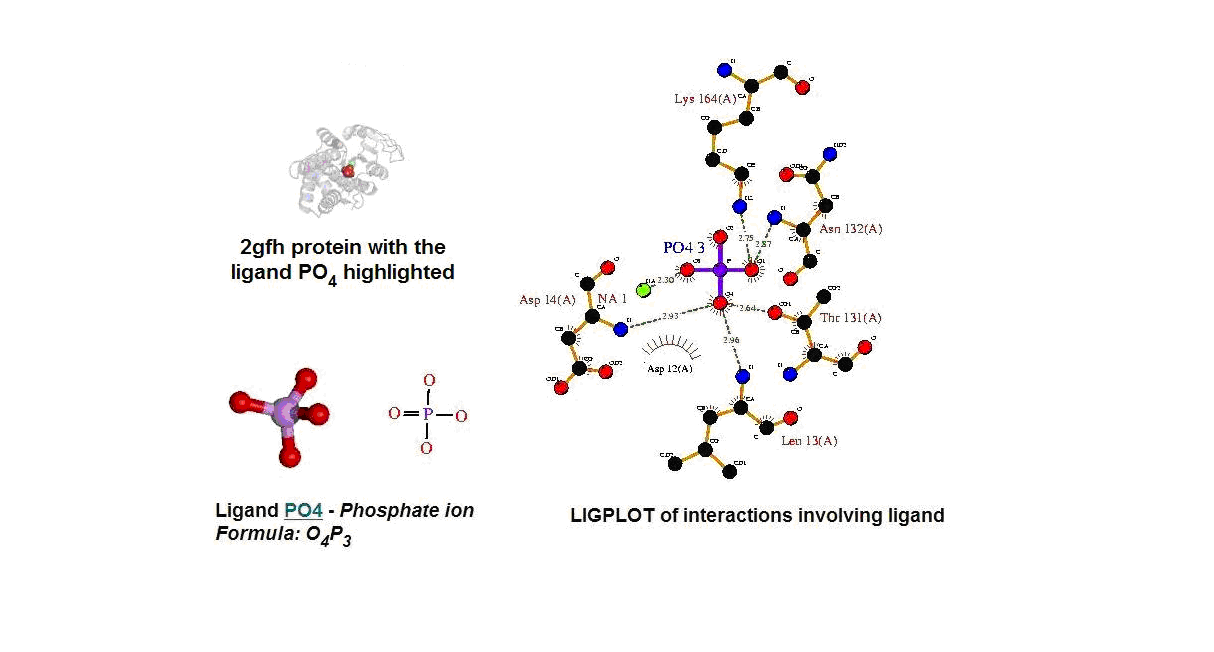 Figure 13. (A) Molecular structure of 2gfh with the ligand PO4. (B) Molecular and chemical structure of PO4. (C) Ligand interaction involving PO4. (Picture adapted from Profunc)
Figure 13. (A) Molecular structure of 2gfh with the ligand PO4. (B) Molecular and chemical structure of PO4. (C) Ligand interaction involving PO4. (Picture adapted from Profunc)
- Identify the likely biochemical function from the 3D structure
- Possible binding sites and potential ligands - PO4
- PO4 most likely be an active site and fuction
Phostphatase
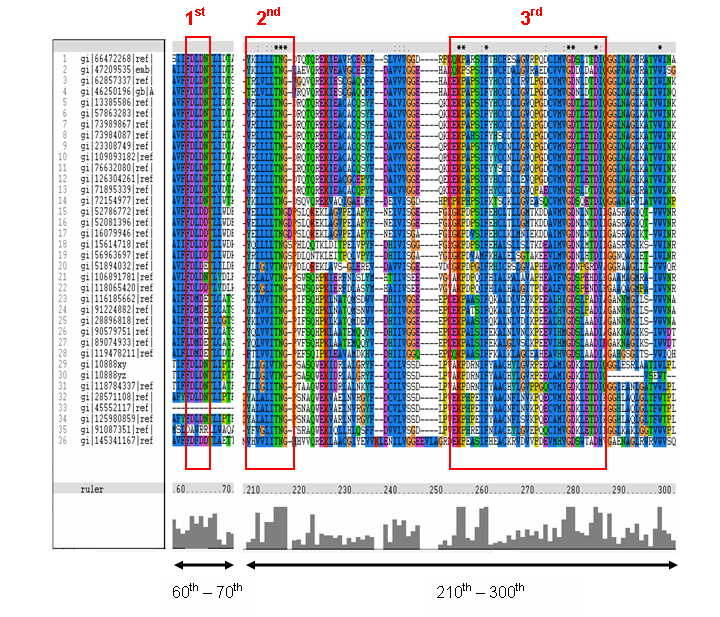
MSA of the 2gfh with 35 others proteins. Only the 60th – 70th and the 210th -300th amino acid
sequence were shown to illustrate the conserved and invariant regions. The 3 boxed-up sequences were either conserved or invariant regions.
- 1st - aspartic acid (D)
- 2nd - threonine (T), asparagine (N) and glycine (G)
- 3rd - lysine (K) and aspartates (D)
- MSA corelate with with study done by Maliekal et al
- N-acetylneuraminic acid phosphatase orthologs shared 3 motifs found in phosphatases
, illustrate that 2gfh is a hydrolase. Profunc searches (Figure 17) on 2gfh also
show that it possesses hydrolase activity. The highest score for Gene Ontology (Figure 17) states it used for metabolism and possesses
phosphoglycolate phosphatase activity. Hydrolyase is an enzyme which catalyzes hydrolysis reaction (Figure 18), which is the addition of the
hydrogen and hydroxyl ions of water to a molecule with its consequent splitting into two or more simpler molecules. Hydrolase is the systematic
name for any enzyme of EC class 3.
Figure 18. Hydrolyase catalyze the hydrosis of the chemical bond between A and B, resulting of 2 simple molecules.
The MSA for the query sequence and the other 35 sequences shows several conserved motifs. The 1st conserved motif
consists of almost invariant region of aspartic acid (D), only the 33rd protein (gi: |45552117|)
showing gap. The 2nd motif shows conserved and invariant of leucine (L), threonine (T), asparagine (N) and glycine (G). The
3rd motif shows 2 invariant amino acid residues of lysine (K), proline (P), valine (V), glycine (G), aspartic acid (D) and
isoleucine (I). This correlates with the study done by Maliekal et al and strongly suggested that the query protein is a phosphatase.
MSA of the query protein Neu5Ac phosphatase with 35 others proteins. Only the 60th – 70th and the
210th -300th amino acid sequence were shown to illustrate the conserved and invariant regions. The 3 boxed-up sequences
were either conserved or invariant regions.
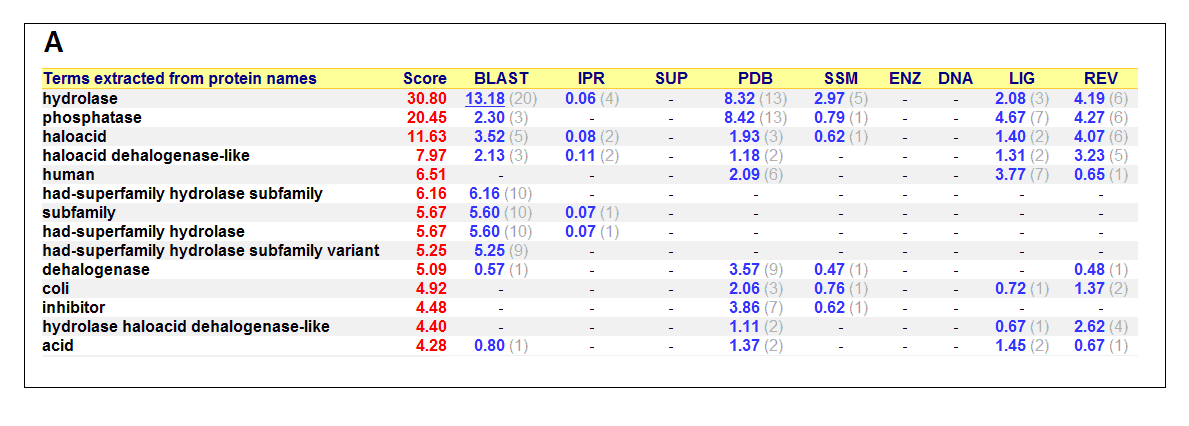
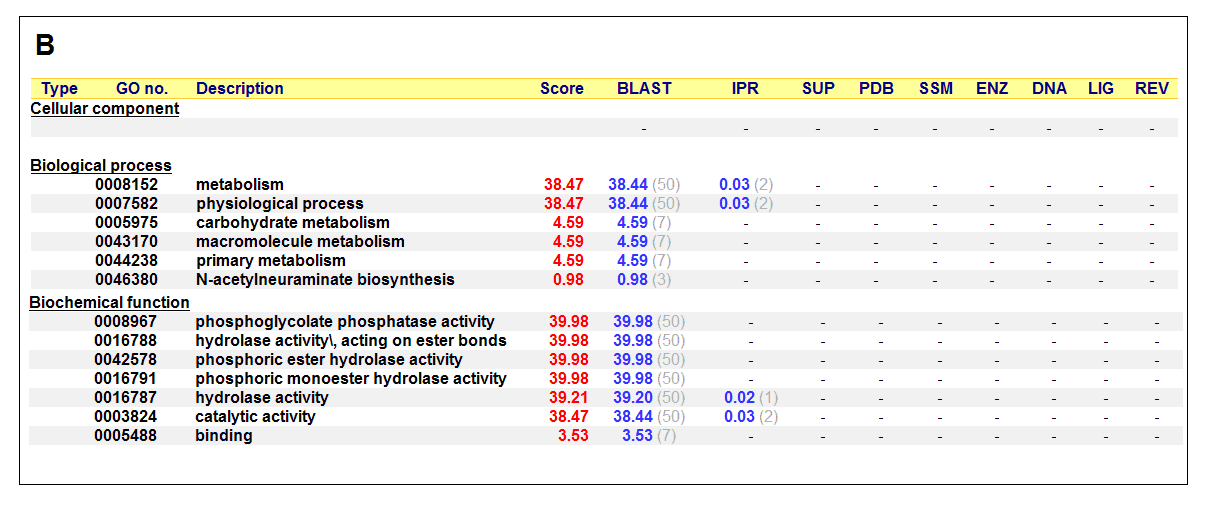 Figure 17. (A) List of all matched protein name terms for 2gfh. (B) List of all matched Gene Ontology terms for 2gfh. The score in
Figure 17. (A) List of all matched protein name terms for 2gfh. (B) List of all matched Gene Ontology terms for 2gfh. The score in
red is a measure of how strongly the term is predicted from the hits obtained by the different methods. The scores in blue show each
method’s contribution to the total score (with the number of relevant sequences/structures shown in brackets in grey).
(http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/)
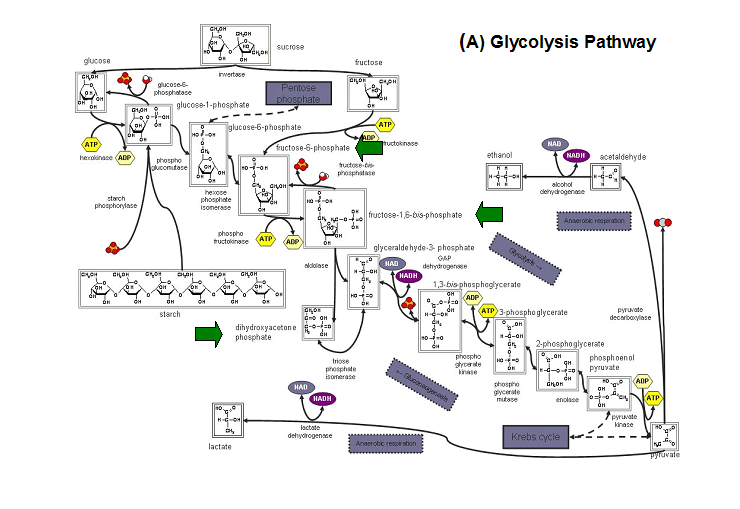
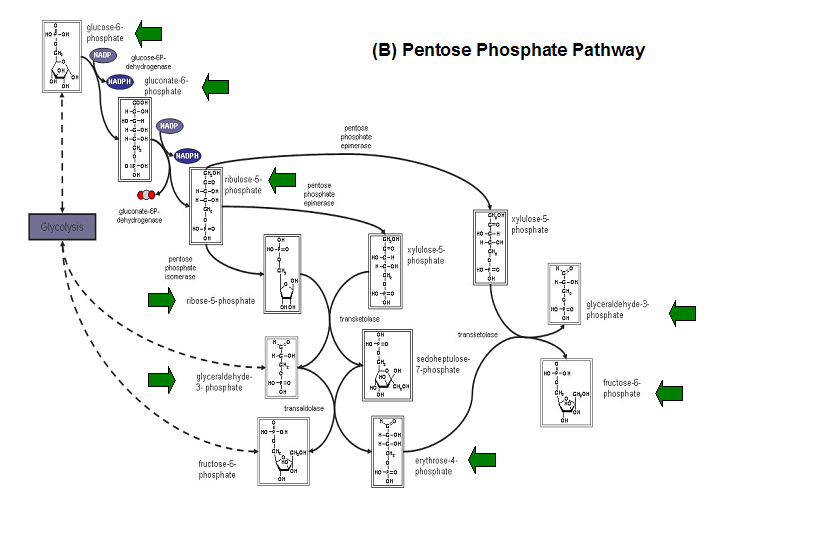
E. coli HADs hydrolyze a wide range of phosphorylated metabolites, including carbohydrates, nucleotides, organic acids, and coenzymes.
Studies have shown that the most common substrates in metabolism such as glycolysis and pentose phosphate pathway (Figure 18). These enzymes
were fructose-1-phosphate, glucose-6-phosphate, mannose-6-phosphate, 2-deoxyglucose-6-phosphate, fructose-6- phosphate, ribose-5-phosphate, and
erythrose- 4-phosphate 13.
Figure 20. The schematic diagrams of glycolysis and pentose phosphate metabolic pathways. The green arrows show the substrates that are hydrolyzed by HADs (A) Glycolysis pathway with substrates that are hydrolyze by HADs: glucose 6-phosphate, fructose 6-phosphate and dihydroxyacetone phosphate. (B) Pentose phostphate pathway with substrates that are hydrolyze by HADs: glucose-6-phosphate, fructose-6-phosphate, dihydroxyacetone phosphate, glyceraldehyde-3-phosphate, gluconate 6-phosphate and erythrose-4-phosphate.
(http://www.steve.gb.com/science/core_metabolism.html)
Proposed Functions
- Hydrolase Activity
- Magnesium Ion Binding
- N-acylneuraminate-9-phosphatase Activity
- Phosphoglycolate Phosphatase Activity
3. N-acylneuraminate-9-phosphatase Activity
4. Phosphoglycolate Phosphatase Activity
| Catalytic activity | N-acylneuraminate 9-phosphate + H2O = N-acylneuraminate + phosphate |
| Cofactor | Magnesium (By similarity) |
| Enzyme regulation | Inhibited by vanadate and calcium (By similarity) |
| Pathway | Carbohydrate metabolism; aminosugar metabolism |
| Similarity | Belongs to the haloacid dehalogenase-like hydrolase superfamily. NANP family |
Using ProFunc: Ligand-binding template search results for 2gfh.
Structural similarity: 91.5%
E-value < 1.00E-06 ( 7.22E-07)
Similarity score: 364.02
PBD id: 2hi0
Name: Hydrolase
Title: Crystal structure of putative phosphoglycolate phosphatase (yp_619066.1) from lactobacillus delbrueckii subsp. Bulgaricus atcc baa-365 at 1.51 a resolution
Source: Lactobacillus delbrueckii. Bacteria. Gene: yp_619066.1. Expressed in: escherichia coli.
Reaction: 2-phosphoglycolate + H2O = glycolate + phosphate
GO Terms
Polymer: haloacid dehalogenase-like hydrolase domain containing 4
Molecular Function: None
Biological Process: None
Cellular Component: None
Function in Human
haloacid dehalogenase-like hydrolase domain
(Found in OMIM)
Neu5Ac-9-PHOSPHATE PHOSPHATASE HALOACID DEHALOGENASE-LIKE HYDROLASE DOMAIN-CONTAINING PROTEIN-4; HDHD4 Gene map locus 20p11
Neu5Ac-9-phosphate phosphatase (Neu5Ac-9-Pase; EC 3.1.3.29) dephosphorylates Neu5Ac-9-phosphate to form N-acetylneuraminate (Neu5Ac), the main form of sialic acid in vertebrates that has important roles in protein-protein and cell-cell recognition.
CLONING
Maliekal et al. (2006) purified Neu5Ac-9-Pase from rat liver and isolated the phosphatase activity. Using SDS-PAGE analysis and tandem mass spectrometry, they identified a haloacid dehalogenase-like hydrolase domain containing-4 (HDHD4) protein. Purified recombinant human HDHD4 catalyzed the dephosphorylation of Neu5Ac-9-phosphate with a catalytic efficiency more than 2 orders of magnitude higher than for any other substrate tested. The 248-amino acid human HDHD4 protein has 3 motifs found in phosphatases of the haloacid dehalogenase (HAD) family: the first with 2 extremely conserved aspartates, the second comprising a conserved serine or threonine, and the third comprising a conserved lysine and 2 conserved aspartates.
GENE STRUCTURE
Maliekal et al. (2006) determined that the human HDHD4 gene contains 2 exons.
GENE FUNCTION
Maliekal et al. (2006) determined that the phosphatase activity of human Neu5Ac-9-Pase protein was dependent on the presence of Mg(2+) and was inhibited by vanadate and Ca(2+), which is characteristic of members of the HAD family of phosphatases.
info
N-acetylneuraminic acid phosphatase
Homologous to mouse (Mus musculus)
Haloacid Dehalogenase Like Hydrolase Domain Containing 4
Classified as Hydrolase
Infomation on ProFunc (useful 2gfhA)
Information on PDBsum (2gfh)
From FASTA most likely function is N-acetylneuraminic acid phosphatase.
The haloalkanoate dehalogenase superfamily (HADSF) is one of the largest and most ubiquitous enzyme families identified to date, with over 3,000 members in organisms ranging from bacteria to humans. Remarkable diversity of chemistry and function has emerged through evolution of the HAD catalytic scaffold. Despite the name, the dehalogenases, which catalyze carbon group transfer, represent a minute fraction of the family. All other known catalytic activities are directed at phosphoryl transfer. Numerous proteins from the HADSF are found in each organism (29 in E. coli and 58 in humans, for example) where they perform a diverse collection of novel physiological functions in primary and secondary metabolism, membrane transport, signal transduction, and nucleic acid repair. http://biophysics.bumc.bu.edu/faculty/allen/allenpage/had.htm
Haloacid dehalogenases (E.C.3.8.1.2) are members of the haloacid dehalogenase superfamily, which also contains ATPases, phosphatases and epoxide hydrolases. They catalyse the conversion of α-halo-carboxylic acids to the corresponding hydroxyalkanoic acid by nucleophilic attack on the α-carbon by a conserved aspartic acid residue to form an ester intermediate, which is then further hydrolysed by a water molecule. There are three subtypes of haloacid dehalogenase based on substrate specificity; those that can use both enantiomers as substrates, those that act only on the L enantiomer and those that act only on the D enantiomer.
Press here to go Back