Arylformamidase Function Slide 4: Difference between revisions
From MDWiki
Jump to navigationJump to search
Thomasparker (talk | contribs) No edit summary |
Thomasparker (talk | contribs) |
||
| Line 1: | Line 1: | ||
== | == Thermostable? == | ||
'''Structural similarity''' | |||
No: Chain Z rmsd lali nres %id Description | No: Chain Z rmsd lali nres %id Description | ||
| Line 9: | Line 9: | ||
14: 1evq-A 21.4 2.9 225 308 18 MOLECULE: SERINE HYDROLASE; | 14: 1evq-A 21.4 2.9 225 308 18 MOLECULE: SERINE HYDROLASE; | ||
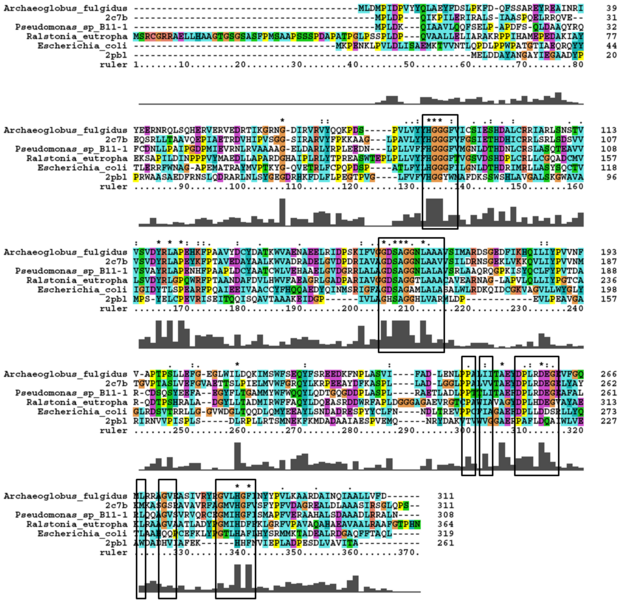
'''Sequence similarity''' | |||
[[Image:HSL_alignment.png|centre|framed|'''Alignment with thermophillic HSL family members identified by Byun 2007.''']] | [[Image:HSL_alignment.png|centre|framed|'''Alignment with thermophillic HSL family members identified by Byun 2007.''']] | ||
[[Arylformamidase Function Slide 3| ...Previous slide ]]|[[Arylformamidase| Return to the main page ]]|[[Arylformamidase Function Slide 5| Next slide... ]] | [[Arylformamidase Function Slide 3| ...Previous slide ]]|[[Arylformamidase| Return to the main page ]]|[[Arylformamidase Function Slide 5| Next slide... ]] | ||
Revision as of 11:38, 3 June 2008
Thermostable?
Structural similarity
No: Chain Z rmsd lali nres %id Description 1: 2pbl-A 52.4 0.0 262 262 100 MOLECULE: PUTATIVE ESTERASE/LIPASE/THIOESTERASE; 5: 2c7b-A 23.4 3.0 231 294 16 MOLECULE: CARBOXYLESTERASE; 9: 1jji-A 23.0 3.1 233 311 15 MOLECULE: CARBOXYLESTERASE; 14: 1evq-A 21.4 2.9 225 308 18 MOLECULE: SERINE HYDROLASE;
Sequence similarity