DAP method: Difference between revisions
TengMengHua (talk | contribs) No edit summary |
No edit summary |
||
| (22 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
== | == BlastP == | ||
: | : | ||
| Line 111: | Line 110: | ||
== | ==ClustalX== | ||
: | : | ||
[[Image:clustalexample.JPG|frame|Multiple Sequence Alignment example]] | [[Image:clustalexample.JPG|frame|'''Figure 1.1''' Multiple Sequence Alignment example]] | ||
Used ClustalX 1.83multiple alignment software tool to align '''C:\3rdplaceoutnames.fasta. '''Output format options was changed to NODE before bootstrapping, this is performed in order see reliability of branches in treeview. | Used ClustalX 1.83multiple alignment software tool to align '''C:\3rdplaceoutnames.fasta. '''Output format options was changed to NODE before bootstrapping, this is performed in order see reliability of branches in treeview. | ||
| Line 128: | Line 127: | ||
===Treeview=== | |||
| Line 139: | Line 138: | ||
[[image:Motif_Sequence1.JPG]] | ==Protein Folding== | ||
: | |||
First DALI search was done to compare the 3D structure with those in the protein data bank. It revealed that Aspartyl Aminopeptidase is a mol1A molecule: Probable M18-Family Aminopeptidase 2. Searching the PDB was then done to source for the structures of biological macromolecules and their relationships to sequence, function, and disease. CE which is a databases and tool for 3-D protein structure ccomparison and alignment was used to compare the alignments between the query protein and its neigbhours. | |||
==Sequence Similarity== | |||
: | |||
Interproscan was then used to analyze the newly determined sequences for annotation of predicted proteins from genome sequencing projects. In order to further analyze the protein, Pfam which is a large collection of multiple sequence alignments and hidden Markov models is used to analyze the protein in this case acetylneuraminic acid phosphatase to find Pfam family matches. The aim of using the ProFunc server is to help identify the likely biochemical function of a protein from its three-dimensional structure. It uses a series of methods, including fold matching, residue conservation, surface cleft analysis, and functional 3D templates, to identify both the protein’s likely active site and possible homologues in the PDB. | |||
==MOTIF identification== | |||
: | |||
MOTIFs were identified using the PROSITE motif search service (Bairoch, Bucher, & Hofmann, 1997) on the Aspartyl Aminopeptidase Chain A residue sequence. The identified MOTIF patterns can be seen below. | |||
[[image:Motif_Sequence1.JPG|framed|<br>'''Figure 1.2''' PROSITE Motif identification of 2ijz Chain A.(Continued in Figure 1.3)|none]]<br> | |||
[[image:Motif_Sequence2.JPG|framed|<br>'''Figure 1.3''' PROSITE Motif Identification of 2ijz Chain A.|none]]<br> | |||
==Structural Alignment== | |||
: | |||
PyMOL was used to align two different sequence structure together to see how closely related they are in a three dimensional diagram. | |||
== EBI-EMBL == | |||
This site is a great resource for finding information on genomics. It can analyse a sequence and had links to many other databases, tools, and journals. | |||
==CluSTr== | |||
Able to provide a link to the data base UniProt and provide a structural alignment of the protein to mouse. | |||
==ExPASy== | |||
'''Prosite''' performed a scan using ProRule | |||
Prosite predicted possible active sites with a high probability of occurence based on sequence data. The output did not take into account enough of the predicted Asp, Glu or His residues to be considered reliable. | |||
[[Image:prositeprediction.jpg|framed|<br>'''Figure 1.4''' Prosite predicted possible active sites with a high probability of occurence based on sequence data. The output did not take into account enough of the predicted Asp, Glu or His residues to be considered reliable.|none]]<br> | |||
==UniProt== | |||
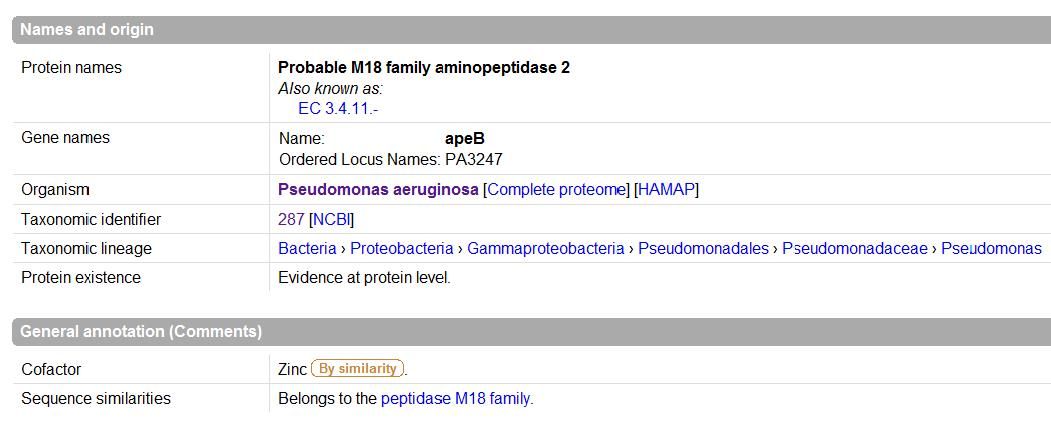
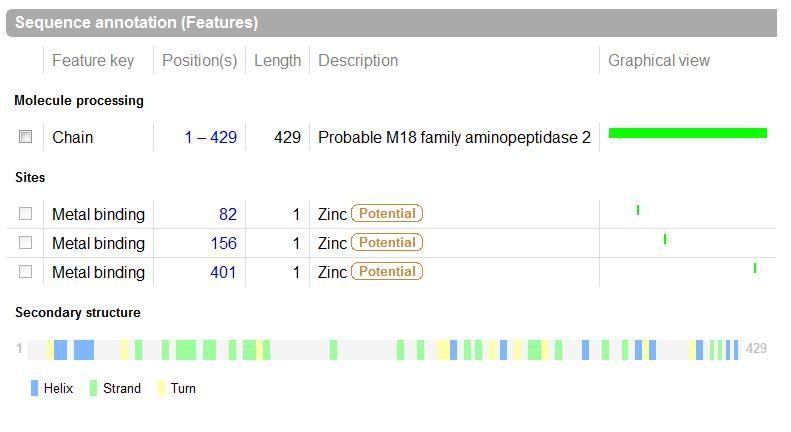
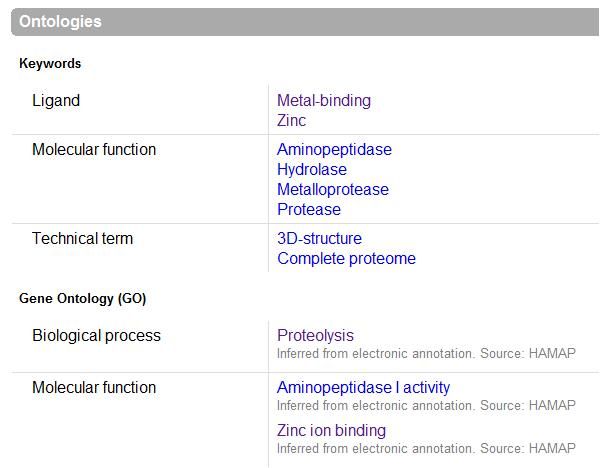
UniProt was used to Identify the function based on sequence in FASTA format, and confirm possible active site residues. | |||
[[Image:names.jpg|framed|<br>'''Figure 1.5''' UniProt output |none]]<br> | |||
[[Image:sequence.jpg|framed|<br>'''Figure 1.6''' UniProt output|none]]<br> | |||
[[Image:ontology.jpg|framed|<br>'''Figure 1.7''' UniProt output|none]]<br> | |||
== Other == | |||
Other Useful resources used are Profunc, pfam, Symatalas which gives expression data, and MEROPS | |||
[http://compbio.chemistry.uq.edu.au/mediawiki/index.php/Aspartyl_Aminopeptidase]Return to Aspartyl Aminopeptidase | |||
Latest revision as of 15:13, 9 June 2008
BlastP
- FASTA SEQUENCE FROM NCBI ENTREZ protein = 2IJZ_A
- Origin of query sequence = Pseudomonas aeruginosa
>gi|119390187|pdb|2IJZ|A Chain A, Crystal Structure Of Aminopeptidase RAELNQGLIDFLKASPTPFHATASLARRLEAAGYRRLDERDAWHTETGGRYYVTRNDSSLIAIRLGRRSP LESGFRLVGAHTDSPCLRVKPNPEIARNGFLQLGVEVYGGALFAPWFDRDLSLAGRVTFRANGKLESR LVDFRKAIAVIPNLNIHLNRAANEGWPINAQNELPPIIAQLAPGEAADFRLLLDEQLLREHGITADVVLDYE LSFYDTQSAAVVGLNDEFIAGARLDNLLSCHAGLEALLNAEGDENCILVCTDHEEVGSCSHCGADGPFLE QVLRRLLPEGDAFSRAIQRSLLVSADNAHGVHPNYADRHDANHGPALNGGPVIKINSNQRYATNSETA GFFRHLCQDSEVPVQSFVTRSDMGCGSTIGPITASQVGVRTVDIGLPTFAMHSIRELAGSHDLAHLVKVLGA FYASSELP
- Performed blastp search against non-redundant (nr) databases which was provided on the CD provided. Query sequences used was Pseudomonas Aeruginosa chain A crystal structure of asparytl aminopeptidase.
- Initial sequence alignment was performed using ClustalX and edited to reduce gapping in the alignment and final multiple sequence alignment was again performed with 38 sequences.
- Treeview32 software was used to view phylogenetic tree produced from multiple sequence alignment and a bootstrapped N-J tree was produced using Clustalx for branches reliability indications.
As mentioned in the methods and website :
C:\blast\blastall -p blastp -d C:\blast\databases\nr -i yourfile.fasta -o usefuloutputname.html
Obtained fastaformat files
C:\blast\fastacmd -d C:\blast\databases\nr -i filewith_img_numbers -o C:\newsequences.fasta
Inputs used for obtaining fastaformat files:
pdb|2IJZ|A ref|YP_789908.1| ref|YP_261475.1| ref|ZP_00416764.1| ref|NP_743887.1| ref|NP_793647.1| ref|YP_607123.1| ref|YP_958321.1| ref|ZP_01894798.1| ref|ZP_01166960.1| ref|ZP_01738318.1| ref|YP_436072.1| ref|ZP_01462550.1| ref|YP_630602.1| ref|YP_001615044.1| ref|YP_747571.1| ref|YP_113441.1| ref|XP_001751765.1| ref|XP_001641062.1| ref|XP_713998.1| gb|AAM61631.1| ref|XP_365906.1| ref|XP_843934.1| ref|NP_001045513.1| ref|XP_001566576.1| ref|XP_001877081.1| gb|ACC64563.1| ref|XP_001492028.1| ref|NP_001039417.1| ref|YP_833603.1| ref|NP_036232.2| ref|NP_001012937.1| ref|NP_001104301.1| gb|EDL75426.1| ref|NP_001085525.1| ref|XP_462175.1| ref|NP_956447.1|
Changed headings in every single obtained fasta sequences into organism names only, e.g :
From
>gi|116051260|ref|YP_789908.1| putative aminopeptidase 2 [Pseudomonas aeruginosa UCBPP-PA14]
MRAELNQGLIDFLKASPTPFHATASLARRLEAAGYRRLDERDAWHTEAGGRYYVTRNDSSLIAIRLGRRSPLESGFRLVG
AHTDSPCLRVKPNPEIARNGFLQLGVEVYGGALFAPWFDRDLSLAGRVTFRANGKLESRLVDFRKAIAVIPNLAIHLNRA
ANEGWPINAQNELPPIIAQLAPGEAADFRLLLDEQLLREHGITADVVLDYELSFYDTQSAAVVGLNDEFIAGARLDNLLS
CHAGLEALLNAEGDENCILVCTDHEEVGSCSHCGADGPFLEQVLRRLLPEGDAFSRAIQRSLLVSADNAHGVHPNYADK
DANHGPALNGGPVIKINSNQRYATNSETAGFFRHLCQDSEVPVQSFVTRSDMGCGSTIGPITASQVGVRTVDIGLPTFAM
HSIRELAGSHDLAHLVKVLGAFYASSELP
To
>Pseudomonas_aeruginosa
MRAELNQGLIDFLKASPTPFHATASLARRLEAAGYRRLDERDAWHTEAGGRYYVTRNDSSLIAIRLGRRSPLESGFRLVG
AHTDSPCLRVKPNPEIARNGFLQLGVEVYGGALFAPWFDRDLSLAGRVTFRANGKLESRLVDFRKAIAVIPNLAIHLNRA
ANEGWPINAQNELPPIIAQLAPGEAADFRLLLDEQLLREHGITADVVLDYELSFYDTQSAAVVGLNDEFIAGARLDNLLS
CHAGLEALLNAEGDENCILVCTDHEEVGSCSHCGADGPFLEQVLRRLLPEGDAFSRAIQRSLLVSADNAHGVHPNYADK
DANHGPALNGGPVIKINSNQRYATNSETAGFFRHLCQDSEVPVQSFVTRSDMGCGSTIGPITASQVGVRTVDIGLPTFAM
HSIRELAGSHDLAHLVKVLGAFYASSELP
saved into a new file organismnames.fasta
ClustalX
Used ClustalX 1.83multiple alignment software tool to align C:\3rdplaceoutnames.fasta. Output format options was changed to NODE before bootstrapping, this is performed in order see reliability of branches in treeview.
Conserved regions (*) of >gi|119390187|pdb|2IJZ|A Chain A, Crystal Structure Of Aminopeptidase was noted for structural analysis.
Output obtained : .aln file (alignment) and .dnd file (output guide tree)
Bootstrapping : .phb file obtained
Treeview
Used Treeview to visualize Phylogenetic tree:
- Radial Tree
- Rectangular Cladogram
The results from the blast search were then screened and a selection was of these results were used for a multiple sequence alignment using ClustalX. This result was boostrapped and these values checked and more sequences were added to improve the resolution of specific branches. A bootstrapped phylogram was produced, as well as a radial tree.
Protein Folding
First DALI search was done to compare the 3D structure with those in the protein data bank. It revealed that Aspartyl Aminopeptidase is a mol1A molecule: Probable M18-Family Aminopeptidase 2. Searching the PDB was then done to source for the structures of biological macromolecules and their relationships to sequence, function, and disease. CE which is a databases and tool for 3-D protein structure ccomparison and alignment was used to compare the alignments between the query protein and its neigbhours.
Sequence Similarity
Interproscan was then used to analyze the newly determined sequences for annotation of predicted proteins from genome sequencing projects. In order to further analyze the protein, Pfam which is a large collection of multiple sequence alignments and hidden Markov models is used to analyze the protein in this case acetylneuraminic acid phosphatase to find Pfam family matches. The aim of using the ProFunc server is to help identify the likely biochemical function of a protein from its three-dimensional structure. It uses a series of methods, including fold matching, residue conservation, surface cleft analysis, and functional 3D templates, to identify both the protein’s likely active site and possible homologues in the PDB.
MOTIF identification
MOTIFs were identified using the PROSITE motif search service (Bairoch, Bucher, & Hofmann, 1997) on the Aspartyl Aminopeptidase Chain A residue sequence. The identified MOTIF patterns can be seen below.
Structural Alignment
PyMOL was used to align two different sequence structure together to see how closely related they are in a three dimensional diagram.
EBI-EMBL
This site is a great resource for finding information on genomics. It can analyse a sequence and had links to many other databases, tools, and journals.
CluSTr
Able to provide a link to the data base UniProt and provide a structural alignment of the protein to mouse.
ExPASy
Prosite performed a scan using ProRule
Prosite predicted possible active sites with a high probability of occurence based on sequence data. The output did not take into account enough of the predicted Asp, Glu or His residues to be considered reliable.
UniProt
UniProt was used to Identify the function based on sequence in FASTA format, and confirm possible active site residues.
Other
Other Useful resources used are Profunc, pfam, Symatalas which gives expression data, and MEROPS
[1]Return to Aspartyl Aminopeptidase