Structure: Difference between revisions
No edit summary |
|||
| (26 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
== | == Materials & Methods== | ||
1) Protein structural search on PyMOL, NCBI Entrez, InterPro and PDB databases | |||
* PDB code: 3b5q | |||
2) Comparing protein search with DALI server to identify similiar structures | |||
== | |||
== Results == | |||
'''PDB''' | |||
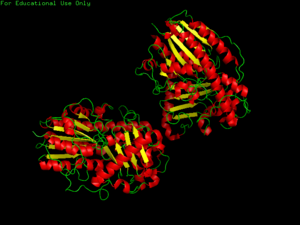
[[Image:3b5q image PDB.png|thumb|'''Figure 1.0''' Structure of 3b5q]] | |||
== Discussion == | |||
'''Sequence''' | |||
>gi|160286517|pdb|3B5Q|A Chain A, Crystal Structure Of A Putative Sulfatase (Np_810509.1) From Bacteroides Thetaiotaomicron Vpi-5482 At 2.40 A Resolution | |||
GXGLALCGAAAQAQEKPNFLIIQCDHLTQRVVGAYGQTQGCTLPIDEVASRGVIFSNAYVGCPLSQPSRA | |||
ALWSGXXPHQTNVRSNSSEPVNTRLPENVPTLGSLFSESGYEAVHFGKTHDXGSLRGFKHKEPVAKPFTD | |||
PEFPVNNDSFLDVGTCEDAVAYLSNPPKEPFICIADFQNPHNICGFIGENAGVHTDRPISGPLPELPDNF | |||
DVEDWSNIPTPVQYICCSHRRXTQAAHWNEENYRHYIAAFQHYTKXVSKQVDSVLKALYSTPAGRNTIVV | |||
IXADHGDGXASHRXVTKHISFYDEXTNVPFIFAGPGIKQQKKPVDHLLTQPTLDLLPTLCDLAGIAVPAE | |||
KAGISLAPTLRGEKQKKSHPYVVSEWHSEYEYVTTPGRXVRGPRYKYTHYLEGNGEELYDXKKDPGERKN | |||
LAKDPKYSKILAEHRALLDDYITRSKDDYRSLKVDADPRCRNHTPGYPSHEGPGAREILKRK | |||
== Links == | |||
[http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&val=160286517 Arylsulfatase sequence] | [http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&val=160286517 Arylsulfatase sequence] | ||
| Line 22: | Line 57: | ||
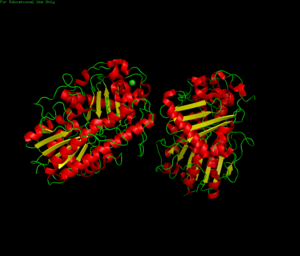
'''Crystal Structure''' | |||
[[Image:protein showing zinc pymol.png|300px|thumb]] | |||
Latest revision as of 01:25, 21 May 2008
Materials & Methods
1) Protein structural search on PyMOL, NCBI Entrez, InterPro and PDB databases
- PDB code: 3b5q
2) Comparing protein search with DALI server to identify similiar structures
Results
PDB
Discussion
Sequence
>gi|160286517|pdb|3B5Q|A Chain A, Crystal Structure Of A Putative Sulfatase (Np_810509.1) From Bacteroides Thetaiotaomicron Vpi-5482 At 2.40 A Resolution GXGLALCGAAAQAQEKPNFLIIQCDHLTQRVVGAYGQTQGCTLPIDEVASRGVIFSNAYVGCPLSQPSRA ALWSGXXPHQTNVRSNSSEPVNTRLPENVPTLGSLFSESGYEAVHFGKTHDXGSLRGFKHKEPVAKPFTD PEFPVNNDSFLDVGTCEDAVAYLSNPPKEPFICIADFQNPHNICGFIGENAGVHTDRPISGPLPELPDNF DVEDWSNIPTPVQYICCSHRRXTQAAHWNEENYRHYIAAFQHYTKXVSKQVDSVLKALYSTPAGRNTIVV IXADHGDGXASHRXVTKHISFYDEXTNVPFIFAGPGIKQQKKPVDHLLTQPTLDLLPTLCDLAGIAVPAE KAGISLAPTLRGEKQKKSHPYVVSEWHSEYEYVTTPGRXVRGPRYKYTHYLEGNGEELYDXKKDPGERKN LAKDPKYSKILAEHRALLDDYITRSKDDYRSLKVDADPRCRNHTPGYPSHEGPGAREILKRK
Links
Human sulfatases: a structural perspective to catalysis
Crystal Structure