Structure: Difference between revisions
From MDWiki
Jump to navigationJump to search
No edit summary |
No edit summary |
||
| Line 10: | Line 10: | ||
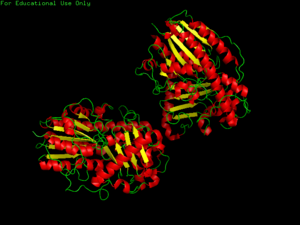
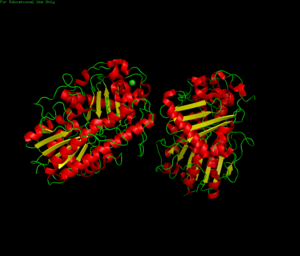
[[Image:3b5q image PDB.png|thumb|'''Figure 1.0''' Structure of 3b5q]] | [[Image:3b5q image PDB.png|thumb|'''Figure 1.0''' Structure of 3b5q]] | ||
Revision as of 10:05, 20 May 2008
Materials & Methods
1) Protein structural search on PyMOL, NCBI Entrez, InterPro and PDB databases
- PDB code: 3b5q
2) Comparing protein search with DALI server to identify similiar structures
Results
PDB
Discussion
Sequence
1 gxglalcgaa aqaqekpnfl iiqcdhltqr vvgaygqtqg ctlpidevas rgvifsnayv
61 gcplsqpsra alwsgxxphq tnvrsnssep vntrlpenvp tlgslfsesg yeavhfgkth
121 dxgslrgfkh kepvakpftd pefpvnndsf ldvgtcedav aylsnppkep ficiadfqnp
181 hnicgfigen agvhtdrpis gplpelpdnf dvedwsnipt pvqyiccshr rxtqaahwne
241 enyrhyiaaf qhytkxvskq vdsvlkalys tpagrntivv ixadhgdgxa shrxvtkhis
301 fydextnvpf ifagpgikqq kkpvdhlltq ptldllptlc dlagiavpae kagislaptl
361 rgekqkkshp yvvsewhsey eyvttpgrxv rgprykythy legngeelyd xkkdpgerkn
421 lakdpkyski laehralldd yitrskddyr slkvdadprc rnhtpgypsh egpgareilk
481 rk
Links
Human sulfatases: a structural perspective to catalysis
Crystal Structure