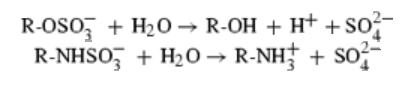3b5q Structure and Function
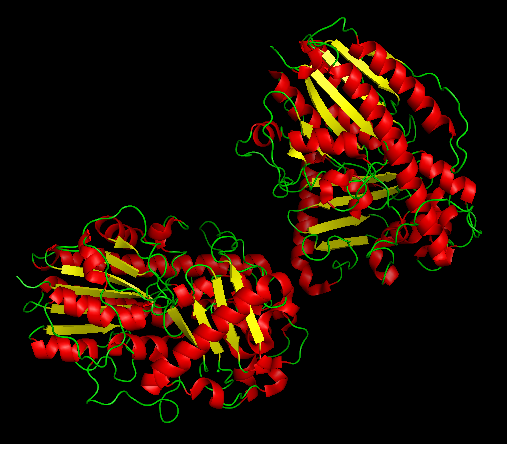
Arylsulfatase K structure
Protein data bank profile characteris arylsulfatase as a hydrolase and a sulfatase. A Sulfatase hydrolyses sulfate esters bonds of substrates, including O-sulfates and N-sulfates as shown below.
Sulfatases are enzymes,which hydrolyse sulfate ester bonds of substrates. These are categorised as EC 3.1.6. in enzyme classifications. Most of the family members has shown to contain a highly conserved cystine residue and a bivalent metal binding site.
Arylsulphatase A (ASA) is a lysosomal enzyme which hydrolyzes cerebroside sulphate. Arylsulphatase B (ASB),also a lysosomal enzyme, which hydrolyzes the sulphate ester group from N-acetylgalactosamine 4-sulphate group of dermatan sulphate.
Sequence conservation
MSA data revealed several highly conserved residues in the N-terminal region of all sequences aligned. These residues are shown below as an alignment of Arylsulfatase K with the sequence of human stroid sulfatase.
Figure 2: Multiple sequence alignment (MSA). Residues which are concerved across the entire sulfatase family are marked in red
Structural alignment
- Three dimentional structure of arylsulfatase was aligned with other available structures using DALI server (structural alignment). Results are shown in 'figure 3'.
No: Chain Z rmsd lali nres %id Description 1: 3b5q-A 73.6 0.0 464 464 100 MOLECULE: PUTATIVE SULFATASE YIDJ; 2: 3b5q-B 70.2 0.3 464 467 100 MOLECULE: PUTATIVE SULFATASE YIDJ; 3: 2qzu-A 35.1 2.5 375 465 25 MOLECULE: PUTATIVE SULFATASE YIDJ; 4: 1fsu 28.7 2.8 344 474 22 MOLECULE: N-ACETYLGALACTOSAMINE-4-SULFATASE; 5: 1n2l-A 28.4 3.0 343 483 25 MOLECULE: ARYLSULFATASE A; 6: 1n2k-A 28.1 3.1 342 482 25 MOLECULE: ARYLSULFATASE A; 7: 1e2s-P 28.1 3.0 341 481 26 MOLECULE: ARYLSULFATASE A; 8: 1e3c-P 28.0 3.1 341 481 26 MOLECULE: ARYLSULFATASE A; 9: 1e33-P 28.0 3.1 342 480 25 MOLECULE: ARYLSULFATASE A; 10: 1e1z-P 27.9 3.0 341 481 26 MOLECULE: ARYLSULFATASE A; 11: 1auk 27.8 3.1 340 481 26 MOLECULE: ARYLSULFATASE A; 12: 1p49-A 27.7 2.9 338 548 24 MOLECULE: STERYL-SULFATASE; 13: 1hdh-B 27.4 3.1 365 525 23 MOLECULE: ARYLSULFATASE; 14: 1hdh-A 27.4 3.0 363 525 23 MOLECULE: ARYLSULFATASE; 15: 2rh6-A 24.3 2.6 257 382 14 MOLECULE: PHOSPHODIESTERASE-NUCLEOTIDE PYROPHOSPHATASE; 16: 2rh6-B 24.2 2.6 257 382 14 MOLECULE: PHOSPHODIESTERASE-NUCLEOTIDE PYROPHOSPHATASE;
Figure 3: Structurally related proteins. (No 1 and 2 are two chains of arylsulfatase K).
- The function of highly related proteins were found using ProFunc
- Arylsulfatase A has both sulfuric ester hydrolase and phosphoric monoester hydrolase activities and localised in lysosomes.
- Steryl-sulfatase is a sulfuric ester hydrolase found in endoplasmin reticulums.
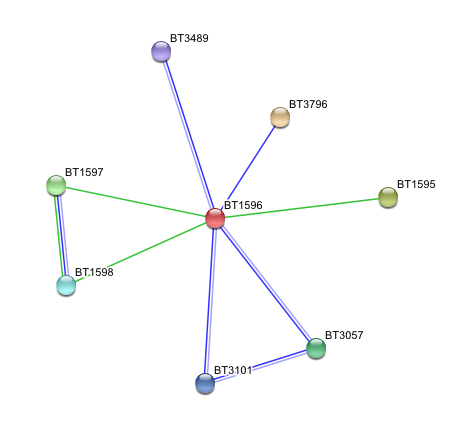
Arylsulfatase K (BT1596) Interactions with other proteins
Input Protein
- BT1596 Putative sulfatase yidJ (481 aa) of Bacteroides thetaiotaomicron.
Predicted Functional Partners
- BT3796: Putative secreted sulfatase ydeN (518 aa).
- BT1595 Transcription termination factor rho (722 aa).
- BT1597 Two-component system sensor histidine kinase (539 aa).
- BT3057 N-acetylgalactosamine-6-sulfatase (508 aa).
- BT1598 Putative two-component system sensor histidine (655 aa).
- BT3101 N-sulphoglucosamine sulphohydrolase (455 aa).
- BT3489 Arylsulfatase B {UniProtKB/TrEMBL-Q8A219} (458 aa).
All these enzymes has generaly the same function, but acts on different substrate.
Finding the functional site and mechanism of action
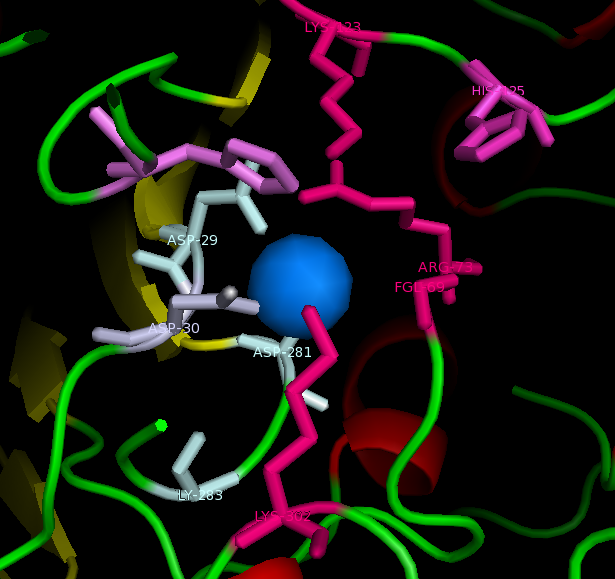
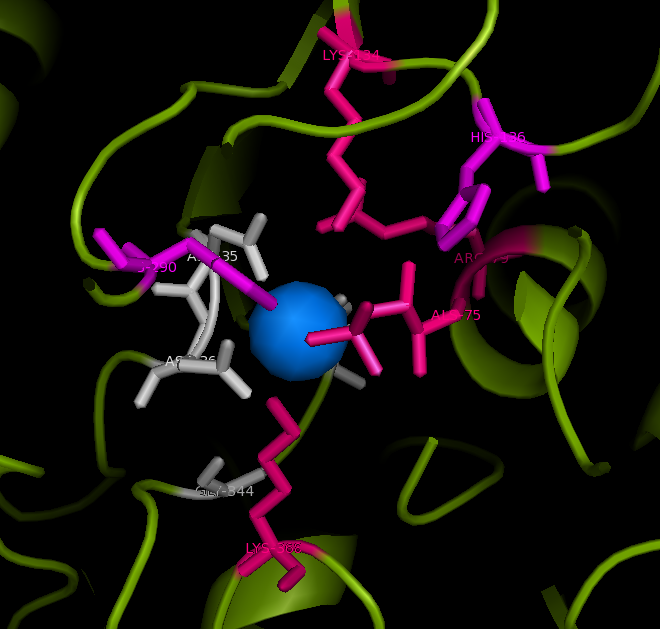
Published data on catalytic activity of STS and ASA was found in literature search. Both STS and ASA shows a divalent metal binding site in the catalytic site, but th crystal structure of ASK doesn't show this. However, three out of four residues which provide oxygen ligans to hold the cation are conserved in ASK. They are shown highlighted in purple in STS-ASK alignment.
The Aspartic acid (D) in 35th position of STS is immidiatly followed by a second aspartic acid. This is substituted by a histidine in ASK. But when the side chain of two residues are examined, it is a possibility for one Nitrogen in five-member-ring to donate a lone pair to form a coordinate bond with a divalent cation. Therefore ASK active site may hold a divalent cation.
Three out of five catalytic resideus were also conserved between ASK and STS (shown highlighted in red). The characteristic cystine residue of sulfatases is conserved in STS, but replaced by serine in ASK. Biologycally acive STS has cystine post translationally modified in to a Formylglycine (FG). The crystal structure doesn't show a FG in ASK structure, however this may be a result of inadequite resolution. The crystal structure has obtained at 2.4 A resolution while bond lenghts of C=O and C-O are 1.2 A and 1.4 A respectively. Further evidence from literature suggests that a subset of ASK found in bacteria including Bacteroides thetaiotamicron and Klebsiella pneumoneae use a S, post translationally modified to a FG, for the same cataltic action[[1]].Finally H290 of STS in not conserven in either ASK or ASA, but the 3D structure shows a different H in the colse proximity and this H in ASA is conserved in most ASA from different species.
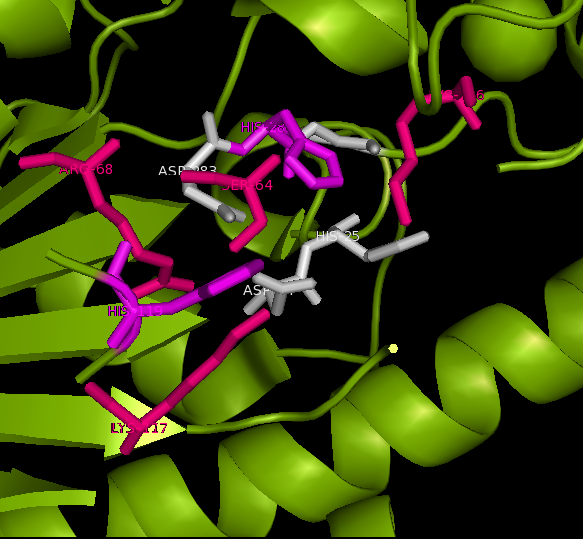
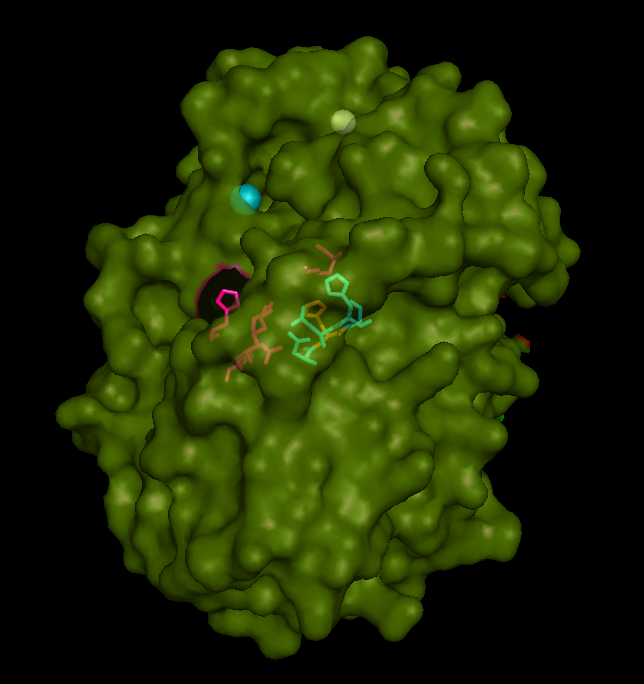
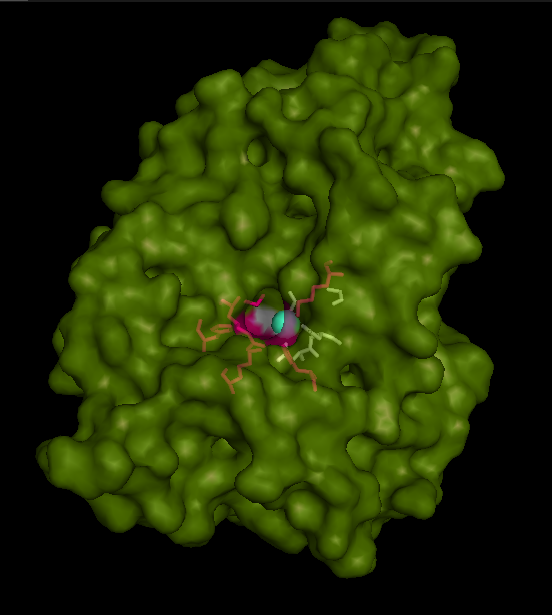
Catalytic site and surface architecture
Possible function and likely substrates of ASK
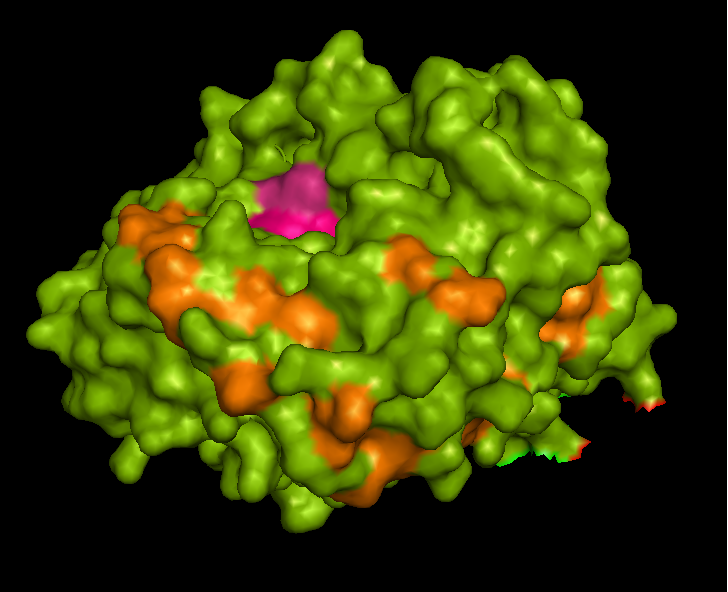
STS is a membrane bound enzyme, mostly found in human placenta and skin fibroblasts. It converts sex-stroid precursors to ative estrogen and androgen, thereofre give a local suppley of these hormones [3] Its catalytic site is buried deep with in the molecule and only connected to the exterior via a substrate-entry path. ASA is a water soluble enzyme found in lysosomes, where the catalytic site is closer to the surface and substrate-entry path is narrower and wider in contrast to that of STS. Structural comparison shows ASK catalsytic site architecture is much similar to that of ASA, eventhough subcellular localization is very different. ASA binds to membrane lipid sulphatides and show a number of hydrophibic patches near the catalytic site. However an exact substrate binding site is not defined. ASK structure also show number of hydrophobic residues on the surface (Figure 11) indication the hydrophobic nature of the substrate it binds.
Conclusion
ASK is a water soluble enzyme localysed in ER. No tissue localization is specified to date. Based on all above evidences the possible function of ASK is hydrolysis of sulphur ester bonds. Considering the architecture and hydrophobicity of surface catalytic residues, ASK may be binding to substrates which are larger in size to stroids that binds STS and hydrophobic in nature such as sulfated lipids.
Click here to go Back